P23440
Gene name |
Pde6b |
Protein name |
Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta |
Names |
GMP-PDE beta |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:18587 |
EC number |
3.1.4.35: Phosphoric diester hydrolases |
Protein Class |
|
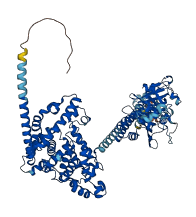
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P23440
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P23440-F1 | Predicted | AlphaFoldDB |
18 variants for P23440
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388771751 | 119 | L>M | No | EVA | |
| rs3388777780 | 142 | V>A | No | EVA | |
| rs3388782890 | 153 | D>H | No | EVA | |
| rs3388780407 | 172 | T>A | No | EVA | |
| rs3388782948 | 223 | Y>N | No | EVA | |
| rs3389582336 | 296 | E>K | No | EVA | |
| rs3396025423 | 305 | T>I | No | EVA | |
| rs3396019888 | 307 | D>A* | No | EVA | |
| rs3388777859 | 316 | V>A | No | EVA | |
| rs3388761705 | 338 | W>* | No | EVA | |
| rs49995481 | 347 | Y>* | No | EVA | |
| rs248731758 | 360 | A>V | No | EVA | |
| rs3388777868 | 407 | G>R | No | EVA | |
| rs3388764716 | 460 | K>N | No | EVA | |
| rs3388775290 | 555 | T>S | No | EVA | |
| rs3388778301 | 678 | M>L | No | EVA | |
| rs3388772925 | 745 | D>N | No | EVA | |
| rs3388780439 | 831 | R>G | No | EVA |
1 associated diseases with P23440
Without disease ID
5 regional properties for P23440
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | 3'5'-cyclic nucleotide phosphodiesterase, catalytic domain | 481 - 814 | IPR002073 |
| domain | GAF domain | 71 - 230 | IPR003018-1 |
| domain | GAF domain | 252 - 439 | IPR003018-2 |
| domain | HD/PDEase domain | 554 - 741 | IPR003607 |
| conserved_site | 3'5'-cyclic nucleotide phosphodiesterase, conserved site | 597 - 608 | IPR023174 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.4.35 | Phosphoric diester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| photoreceptor outer segment | The outer segment of a vertebrate photoreceptor that contains a stack of membrane discs embedded with photoreceptor proteins. |
| photoreceptor outer segment membrane | The membrane surrounding the outer segment of a vertebrate photoreceptor. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| 3',5'-cyclic-GMP phosphodiesterase activity | Catalysis of the reaction: 3',5'-cyclic GMP + H2O = GMP + H+. |
| 3',5'-cyclic-nucleotide phosphodiesterase activity | Catalysis of the reaction: a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate. |
| metal ion binding | Binding to a metal ion. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| detection of light stimulus | The series of events in which a light stimulus (in the form of photons) is received and converted into a molecular signal. |
| entrainment of circadian clock by photoperiod | The synchronization of a circadian rhythm to photoperiod, the intermittent cycle of light (day) and dark (night). |
| retina development in camera-type eye | The process whose specific outcome is the progression of the retina over time, from its formation to the mature structure. The retina is the innermost layer or coating at the back of the eyeball, which is sensitive to light and in which the optic nerve terminates. |
| retinal cell apoptotic process | Any apoptotic process in a retinal cell. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| visual perception | The series of events required for an organism to receive a visual stimulus, convert it to a molecular signal, and recognize and characterize the signal. Visual stimuli are detected in the form of photons and are processed to form an image. |
33 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q28156 | PDE5A | cGMP-specific 3',5'-cyclic phosphodiesterase | Bos taurus (Bovine) | SS |
| P23439 | PDE6B | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Bos taurus (Bovine) | PR |
| P52731 | PDE6C | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' | Gallus gallus (Chicken) | PR |
| H2QL32 | PDE9A | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Pan troglodytes (Chimpanzee) | PR |
| P33726 | PDE6B | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| Q9W4T4 | dnc | 3',5'-cyclic-AMP phosphodiesterase, isoform I | Drosophila melanogaster (Fruit fly) | SS |
| Q9VJ79 | Pde11 | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11 | Drosophila melanogaster (Fruit fly) | SS |
| Q9HCR9 | PDE11A | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Homo sapiens (Human) | SS |
| O76083 | PDE9A | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Homo sapiens (Human) | PR |
| Q07343 | PDE4B | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | Homo sapiens (Human) | EV SS |
| Q08499 | PDE4D | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | Homo sapiens (Human) | EV |
| O76074 | PDE5A | cGMP-specific 3',5'-cyclic phosphodiesterase | Homo sapiens (Human) | EV |
| Q08493 | PDE4C | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Homo sapiens (Human) | EV SS |
| P51160 | PDE6C | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' | Homo sapiens (Human) | PR |
| P27815 | PDE4A | cAMP-specific 3',5'-cyclic phosphodiesterase 4A | Homo sapiens (Human) | EV SS |
| Q9Y233 | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Homo sapiens (Human) | PR |
| P35913 | PDE6B | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Homo sapiens (Human) | PR |
| O89084 | Pde4a | 3',5'-cyclic-AMP phosphodiesterase 4A | Mus musculus (Mouse) | SS |
| Q3UEI1 | Pde4c | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Mus musculus (Mouse) | PR |
| Q01063 | Pde4d | 3',5'-cyclic-AMP phosphodiesterase 4D | Mus musculus (Mouse) | SS |
| O70628 | Pde9a | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Mus musculus (Mouse) | PR |
| Q8CA95 | Pde10a | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Mus musculus (Mouse) | PR |
| P0C1Q2 | Pde11a | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Mus musculus (Mouse) | SS |
| Q8CG03 | Pde5a | cGMP-specific 3',5'-cyclic phosphodiesterase | Mus musculus (Mouse) | SS |
| Q8QZV1 | Pde9a | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Rattus norvegicus (Rat) | PR |
| P14646 | Pde4b | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | Rattus norvegicus (Rat) | SS |
| Q8VID6 | Pde11a | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Rattus norvegicus (Rat) | SS |
| O54735 | Pde5a | cGMP-specific 3',5'-cyclic phosphodiesterase | Rattus norvegicus (Rat) | SS |
| P14644 | Pde4c | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Rattus norvegicus (Rat) | PR |
| P54748 | Pde4a | 3',5'-cyclic-AMP phosphodiesterase 4A | Rattus norvegicus (Rat) | SS |
| Q9QYJ6 | Pde10a | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Rattus norvegicus (Rat) | PR |
| P14270 | Pde4d | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | Rattus norvegicus (Rat) | PR |
| Q22000 | pde-4 | Probable 3',5'-cyclic phosphodiesterase pde-4 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSLSEEQVRS | FLDGNPTFAH | QYFGKKLSPE | NVAGACEDGW | LADCGSLREL | CQVEESAALF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ELVQDMQESV | NMERVVFKIL | RRLCTILHAD | RCSLFMYRQR | NGIAELATRL | FSVQPDSLLE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DCLVPPDSEI | VFPLDIGIVG | HVAQTKKMIN | VQDVAECPHF | SSFADELTDY | VTKNILSTPI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MNGKDVVAVI | MAVNKLDGPC | FTSEDEDVFT | KYLNFATLNL | KIYHLSYLHN | CETRRGQVLL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| WSANKVFEEL | TDIERQFHKA | FYTVRAYLNC | ERYSVGLLDM | TKEKEFFDVW | PVLMGEAQPY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SGPRTPDGRE | IVFYKVIDYI | LHGKEDIKVI | PTPPADHWAL | ASGLPTYVAE | SGFICNIMNA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SADEMFNFQE | GPLDDSGWVI | KNVLSMPIVN | KKEEIVGVAT | FYNRKDGKPF | DDQDEVLMES |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LTQFLGWSVL | NTDTYDKMNK | LENRKDIAQD | MVLYHVRCDK | DEIQEILPTR | DRLGKEPADC |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EEDELGKILK | EELPGPTKFD | IYEFHFSDLE | CTELELVKCG | IQMYYELGVV | RKFQIPQEVL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VRFLFSVSKA | YRRITYHNWR | HGFNVAQTMF | TLLMTGKLKS | YYTDLEAFAM | VTAGLCHDID |
| 610 | 620 | 630 | 640 | 650 | 660 |
| HRGTNNLYQM | KSQNPLAKLH | GSSILERHHL | EFGKFLLAEE | SLNIYQNLNR | RQHEHVIHLM |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DIAIIATDLA | LYFKKRTMFQ | KIVDESKNYE | DKKSWVEYLS | LETTRKEIVM | AMMMTACDLS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| AITKPWEVQS | KVALLVAAEF | WEQGDLERTV | LDQQPIPMMD | RNKAAELPKL | QVGFIDFVCT |
| 790 | 800 | 810 | 820 | 830 | 840 |
| FVYKEFSRFH | EEILPMFDRL | QNNRKEWKAL | ADEYEAKVKA | LEEEKKKEED | RVAAKKVGTE |
| 850 | |||||
| VCNGGPAPKS | STCCIL |