P23298
Gene name |
Prkch (Pkch) |
Protein name |
Protein kinase C eta type |
Names |
PKC-L, nPKC-eta |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:18755 |
EC number |
2.7.11.13: Protein-serine/threonine kinases |
Protein Class |
|
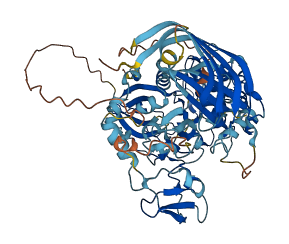
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
496-519 (Activation loop from InterPro)
Target domain |
359-681 (Catalytic domain of the Serine/Threonine Kinase, Novel Protein Kinase C eta) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for P23298
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P23298-F1 | Predicted | AlphaFoldDB |
29 variants for P23298
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389213088 | 102 | T>M | No | EVA | |
| rs3389253077 | 130 | V>M | No | EVA | |
| rs264564113 | 153 | H>N | No | EVA | |
| rs3389243903 | 154 | F>Y | No | EVA | |
| rs3389203553 | 245 | P>T | No | EVA | |
| rs3389253092 | 255 | V>M | No | EVA | |
| rs3389245084 | 256 | P>T | No | EVA | |
| rs3389229806 | 259 | C>S | No | EVA | |
| rs3403752776 | 260 | D>V | No | EVA | |
| rs3389242623 | 270 | M>K | No | EVA | |
| rs3389243873 | 315 | N>I | No | EVA | |
| rs3403218793 | 323 | I>L | No | EVA | |
| rs3403413514 | 330 | R>W | No | EVA | |
| rs3389253150 | 353 | D>V | No | EVA | |
| rs237019456 | 356 | E>D | No | EVA | |
| rs3389239866 | 368 | K>E | No | EVA | |
| rs3389203567 | 385 | V>E | No | EVA | |
| rs3389175924 | 446 | Q>P | No | EVA | |
| rs3412177960 | 454 | A>V | No | EVA | |
| rs3403611758 | 460 | A>D | No | EVA | |
| rs3389242647 | 469 | F>I | No | EVA | |
| rs3389213053 | 503 | E>K | No | EVA | |
| rs3389217160 | 534 | A>P | No | EVA | |
| rs3389257522 | 541 | G>S | No | EVA | |
| rs3389203616 | 562 | L>P | No | EVA | |
| rs3389252296 | 565 | A>G | No | EVA | |
| rs3389246317 | 614 | F>Y | No | EVA | |
| rs3389217115 | 632 | R>P | No | EVA | |
| rs3389175899 | 641 | V>I | No | EVA |
No associated diseases with P23298
8 regional properties for P23298
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Armadillo | 116 - 242 | IPR000225-1 |
| repeat | Armadillo | 245 - 284 | IPR000225-2 |
| repeat | Armadillo | 286 - 326 | IPR000225-3 |
| repeat | Armadillo | 328 - 368 | IPR000225-4 |
| repeat | Armadillo | 370 - 410 | IPR000225-5 |
| repeat | Armadillo | 413 - 453 | IPR000225-6 |
| domain | Importin-alpha, importin-beta-binding domain | 1 - 105 | IPR002652 |
| repeat | Atypical Arm repeat | 467 - 516 | IPR032413 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.13 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
12 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium-dependent protein kinase C activity | Calcium-dependent catalysis of the reaction: ATP + a protein = ADP + a phosphoprotein. |
| calcium-independent protein kinase C activity | Catalysis of the reaction: ATP + a protein = ADP + a phosphoprotein. This reaction requires diacylglycerol but not calcium. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine kinase binding | Binding to a protein serine/threonine kinase. |
| protein serine/threonine kinase inhibitor activity | Binds to and stops, prevents or reduces the activity of a protein serine/threonine kinase. |
| small GTPase binding | Binding to a small monomeric GTPase. |
| translation regulator activity | Any molecular function involved in the initiation, activation, perpetuation, repression or termination of polypeptide synthesis at the ribosome. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cellular response to amino acid starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of amino acids. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of glial cell apoptotic process | Any process that stops, prevents, or reduces the frequency, rate, or extent of glial cell apoptotic process. |
| negative regulation of protein serine/threonine kinase activity | Any process that decreases the rate, frequency, or extent of protein serine/threonine kinase activity. |
| negative regulation of translation | Any process that stops, prevents, or reduces the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of B cell receptor signaling pathway | Any process that activates or increases the frequency, rate or extent of signaling pathways initiated by the cross-linking of an antigen receptor on a B cell. |
| positive regulation of glial cell proliferation | Any process that activates or increases the rate or extent of glial cell proliferation. |
| positive regulation of keratinocyte differentiation | Any process that activates or increases the frequency, rate or extent of keratinocyte differentiation. |
| positive regulation of macrophage derived foam cell differentiation | Any process that increases the rate, frequency or extent of macrophage derived foam cell differentiation. Macrophage derived foam cell differentiation is the process in which a macrophage acquires the specialized features of a foam cell. A foam cell is a type of cell containing lipids in small vacuoles and typically seen in atherosclerotic lesions, as well as other conditions. |
| positive regulation of NF-kappaB transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of the transcription factor NF-kappaB. |
| positive regulation of translation in response to stress | Any process that activates or increases the frequency, rate or extent of translation as a result of a stimulus indicating the organism is under stress. |
| protein kinase C signaling | A series of reactions, mediated by the intracellular serine/threonine kinase protein kinase C, which occurs as a result of a single trigger reaction or compound. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of bicellular tight junction assembly | Any process that modulates the frequency, rate or extent of tight junction assembly. |
47 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P24583 | PKC1 | Protein kinase C-like 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| A1A4I4 | PKN1 | Serine/threonine-protein kinase N1 | Bos taurus (Bovine) | SS |
| P05126 | PRKCB | Protein kinase C beta type | Bos taurus (Bovine) | SS |
| A1Z7T0 | Pkn | Serine/threonine-protein kinase N | Drosophila melanogaster (Fruit fly) | SS |
| P83099 | Pkcdelta | Putative protein kinase C delta type homolog | Drosophila melanogaster (Fruit fly) | PR |
| P05130 | Pkc53E | Protein kinase C, brain isozyme | Drosophila melanogaster (Fruit fly) | SS |
| P13677 | inaC | Protein kinase C, eye isozyme | Drosophila melanogaster (Fruit fly) | SS |
| Q04759 | PRKCQ | Protein kinase C theta type | Homo sapiens (Human) | PR |
| Q6P5Z2 | PKN3 | Serine/threonine-protein kinase N3 | Homo sapiens (Human) | SS |
| Q96LW2 | RSKR | Ribosomal protein S6 kinase-related protein | Homo sapiens (Human) | PR |
| Q16512 | PKN1 | Serine/threonine-protein kinase N1 | Homo sapiens (Human) | EV |
| Q16513 | PKN2 | Serine/threonine-protein kinase N2 | Homo sapiens (Human) | EV |
| Q05655 | PRKCD | Protein kinase C delta type | Homo sapiens (Human) | SS |
| Q02156 | PRKCE | Protein kinase C epsilon type | Homo sapiens (Human) | SS |
| P24723 | PRKCH | Protein kinase C eta type | Homo sapiens (Human) | SS |
| P05129 | PRKCG | Protein kinase C gamma type | Homo sapiens (Human) | SS |
| P17252 | PRKCA | Protein kinase C alpha type | Homo sapiens (Human) | EV |
| P05771 | PRKCB | Protein kinase C beta type | Homo sapiens (Human) | SS |
| P16054 | Prkce | Protein kinase C epsilon type | Mus musculus (Mouse) | PR |
| Q02111 | Prkcq | Protein kinase C theta type | Mus musculus (Mouse) | PR |
| Q8K045 | Pkn3 | Serine/threonine-protein kinase N3 | Mus musculus (Mouse) | SS |
| Q9Z2A0 | Pdpk1 | 3-phosphoinositide-dependent protein kinase 1 | Mus musculus (Mouse) | SS |
| Q02956 | Prkcz | Protein kinase C zeta type | Mus musculus (Mouse) | SS |
| Q62074 | Prkci | Protein kinase C iota type | Mus musculus (Mouse) | SS |
| P70268 | Pkn1 | Serine/threonine-protein kinase N1 | Mus musculus (Mouse) | SS |
| Q8BWW9 | Pkn2 | Serine/threonine-protein kinase N2 | Mus musculus (Mouse) | SS |
| P28867 | Prkcd | Protein kinase C delta type | Mus musculus (Mouse) | PR |
| P20444 | Prkca | Protein kinase C alpha type | Mus musculus (Mouse) | SS |
| P63318 | Prkcg | Protein kinase C gamma type | Mus musculus (Mouse) | SS |
| P68404 | Prkcb | Protein kinase C beta type | Mus musculus (Mouse) | SS |
| Q60823 | Akt2 | RAC-beta serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| P31750 | Akt1 | RAC-alpha serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q9WUA6 | Akt3 | RAC-gamma serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q9ERE3 | Sgk3 | Serine/threonine-protein kinase Sgk3 | Mus musculus (Mouse) | PR |
| Q8QZV4 | Stk32c | Serine/threonine-protein kinase 32C | Mus musculus (Mouse) | PR |
| Q91VJ4 | Stk38 | Serine/threonine-protein kinase 38 | Mus musculus (Mouse) | SS |
| Q7TSE6 | Stk38l | Serine/threonine-protein kinase 38-like | Mus musculus (Mouse) | SS |
| Q63433 | Pkn1 | Serine/threonine-protein kinase N1 | Rattus norvegicus (Rat) | SS |
| P09216 | Prkce | Protein kinase C epsilon type | Rattus norvegicus (Rat) | PR |
| P09215 | Prkcd | Protein kinase C delta type | Rattus norvegicus (Rat) | PR |
| O08874 | Pkn2 | Serine/threonine-protein kinase N2 | Rattus norvegicus (Rat) | SS |
| Q64617 | Prkch | Protein kinase C eta type | Rattus norvegicus (Rat) | PR |
| P63319 | Prkcg | Protein kinase C gamma type | Rattus norvegicus (Rat) | SS |
| P34722 | tpa-1 | Protein kinase C-like 1 | Caenorhabditis elegans | PR |
| P90980 | pkc-2 | Protein kinase C-like 2 | Caenorhabditis elegans | SS |
| A7MBL8 | pkn2 | Serine/threonine-protein kinase N2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q7SY24 | prkcbb | Protein kinase C beta type | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSSGTMKFNG | YLRVRIGEAV | GLQPTRWSLR | HSLFKKGHQL | LDPYLTVSVD | QVRVGQTSTK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QKTNKPTYNE | EFCANVTDGG | HLELAVFHET | PLGYDHFVAN | CTLQFQELLR | TAGTSDTFEG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| WVDLEPEGKV | FVVITLTGSF | TEATLQRDRI | FKHFTRKRQR | AMRRRVHQVN | GHKFMATYLR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QPTYCSHCRE | FIWGVFGKQG | YQCQVCTCVV | HKRCHHLIVT | ACTCQNNINK | VDAKIAEQRF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GINIPHKFNV | HNYKVPTFCD | HCGSLLWGIM | RQGLQCKICK | MNVHIRCQAN | VAPNCGVNAV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ELAKTLAGMG | LQPGNISPTS | KLISRSTLRR | QGKEGSKEGN | GIGVNSSSRF | GIDNFEFIRV |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LGKGSFGKVM | LARIKETGEL | YAVKVLKKDV | ILQDDDVECT | MTEKRILSLA | RNHPFLTQLF |
| 430 | 440 | 450 | 460 | 470 | 480 |
| CCFQTPDRLF | FVMEFVNGGD | LMFHIQKSRR | FDEARARFYA | AEIISALMFL | HEKGIIYRDL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KLDNVLLDHE | GHCKLADFGM | CKEGICNGVT | TATFCGTPDY | IAPEILQEML | YGPAVDWWAM |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GVLLYEMLCG | HAPFEAENED | DLFEAILNDE | VVYPTWLHED | ATGILKSFMT | KNPTMRLGSL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| TQGGEHEILR | HPFFKEIDWA | QLNHRQLEPP | FRPRIKSRED | VSNFDPDFIK | EEPVLTPIDE |
| 670 | 680 | ||||
| GHLPMINQDE | FRNFSYVSPE | LQL |