P22670
Gene name |
RFX1 |
Protein name |
MHC class II regulatory factor RFX1 |
Names |
Enhancer factor C, EF-C, Regulatory factor X 1, RFX, Transcription factor RFX1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5989 |
EC number |
|
Protein Class |
RFX TRANSCRIPTION FACTOR FAMILY (PTHR12619) |
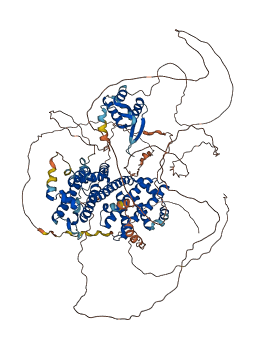
Descriptions
RFX1 is a regulatory factor essential for MHC class II genes expression, and binds to the X boxes of MHC class II genes. This protein possesses intrinsically disordered regions (IDRs) with large negative charge, some of which involve a consecutive sequence of aspartate (D) or glutamate (E) residues, known as D/E repeats. These D/E repeats can cause autoinhibition through intramolecular electrostatic interaction with HMG boxes and modulate binding to DNA. This autoinhibited state can transition into the uninhibited complex with DNA through an electrostatically driven induced-fit process, which accelerates the target DNA search kinetics in the presence of non-functional high-affinity ligands ('decoys').
Autoinhibitory domains (AIDs)
Target domain |
351-527 (DNA binding domain) |
Relief mechanism |
PTM |
Assay |
Deletion assay, Structural analysis |
Accessory elements
No accessory elements
References
- Wang X et al. (2023) "Negatively charged, intrinsically disordered regions can accelerate target search by DNA-binding proteins", Nucleic acids research, 51, 4701-4712
- Katan-Khaykovich Y et al. (2001) "Nuclear import and DNA-binding activity of RFX1. Evidence for an autoinhibitory mechanism", European journal of biochemistry, 268, 3108-16
- Stott K et al. (2010) "Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains", Journal of molecular biology, 403, 706-22
- Watson M et al. (2007) "Mapping intramolecular interactions between domains in HMGB1 using a tail-truncation approach", Journal of molecular biology, 374, 1286-97
- Stott K et al. (2014) "Structural insights into the mechanism of negative regulation of single-box high mobility group proteins by the acidic tail domain", The Journal of biological chemistry, 289, 29817-26
- Ueshima S et al. (2017) "Internal Associations of the Acidic Region of Upstream Binding Factor Control Its Nucleolar Localization", Molecular and cellular biology, 37,
- Wang X et al. (2021) "Dynamic Autoinhibition of the HMGB1 Protein via Electrostatic Fuzzy Interactions of Intrinsically Disordered Regions", Journal of molecular biology, 433, 167122
- Gajiwala KS et al. (2000) "Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding", Nature, 403, 916-21
Autoinhibited structure

Activated structure
2 structures for P22670
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1DP7 | X-ray | 150 A | P | 438-513 | PDB |
| AF-P22670-F1 | Predicted | AlphaFoldDB |
824 variants for P22670
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1447945059 | 3 | T>A | No |
TOPMed gnomAD |
|
| rs752766233 | 4 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1189334998 | 5 | A>E | No |
TOPMed gnomAD |
|
| rs1189334998 | 5 | A>V | No |
TOPMed gnomAD |
|
| rs754885635 | 6 | Y>S | No |
ExAC gnomAD |
|
| rs760268811 | 8 | E>K | No |
ExAC gnomAD |
|
| rs750034252 | 9 | L>P | No |
ExAC gnomAD |
|
| rs767143161 | 10 | Q>K | No |
ExAC gnomAD |
|
| rs761483015 | 10 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs557323175 | 11 | A>V | No |
TOPMed gnomAD |
|
| rs1291276188 | 13 | P>L | No | gnomAD | |
| rs1350376624 | 15 | P>Q | No | gnomAD | |
| rs189912016 | 16 | S>A | No |
1000Genomes gnomAD |
|
| rs189912016 | 16 | S>P | No |
1000Genomes gnomAD |
|
| rs760061480 | 18 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs760061480 | 18 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs747196120 | 19 | P>A | No |
ExAC gnomAD |
|
| rs747196120 | 19 | P>S | No |
ExAC gnomAD |
|
| rs1568480402 | 20 | Q>R | No | Ensembl | |
| rs778033425 | 21 | A>P | No |
ExAC gnomAD |
|
| rs1423239730 | 21 | A>V | No | gnomAD | |
| rs772264878 | 22 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs778930782 | 23 | P>R | No | ExAC | |
| rs1484201925 | 26 | Q>P | No | gnomAD | |
| rs1484201925 | 26 | Q>R | No | gnomAD | |
| rs753759248 | 27 | P>A | No |
ExAC gnomAD |
|
| rs779727703 | 27 | P>H | No |
ExAC gnomAD |
|
| rs779727703 | 27 | P>L | No |
ExAC gnomAD |
|
| rs1473736908 | 28 | Q>P | No | TOPMed | |
| rs905859085 | 29 | P>L | No |
TOPMed gnomAD |
|
| rs1368256370 | 30 | P>R | No | gnomAD | |
| rs1220406116 | 30 | P>S | No | gnomAD | |
| rs371529473 | 31 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1372244924 | 32 | P>L | No | gnomAD | |
| rs761538214 | 33 | P>S | No |
ExAC gnomAD |
|
| rs751158121 | 34 | P>A | No |
ExAC gnomAD |
|
| rs1178347338 | 34 | P>H | No | gnomAD | |
| rs763661107 | 36 | A>V | No |
ExAC gnomAD |
|
| rs777124898 | 37 | A>S | No |
ExAC gnomAD |
|
| rs1473851184 | 38 | P>A | No | gnomAD | |
| rs1473851184 | 38 | P>S | No | gnomAD | |
| rs1200217857 | 39 | Q>P | No |
TOPMed gnomAD |
|
| rs547715748 | 41 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs547715748 | 41 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs868274959 | 41 | P>S | No | Ensembl | |
| rs1599513946 | 42 | Q>P | No | Ensembl | |
| rs532643801 | 43 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs532643801 | 43 | P>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1323547000 | 43 | P>S | No | gnomAD | |
| rs1599513899 | 45 | T>P | No | Ensembl | |
| rs774435556 | 46 | A>S | No |
ExAC gnomAD |
|
|
rs774435556 COSM991790 |
46 | A>T | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs550071125 | 46 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM4074738 | 47 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780108403 | 49 | T>A | No |
ExAC gnomAD |
|
| rs1185358879 | 50 | P>L | No | TOPMed | |
| rs1568480153 | 50 | P>S | No | Ensembl | |
| rs1388073147 | 51 | Q>H | No | gnomAD | |
| rs1599513803 | 51 | Q>P | No | Ensembl | |
| rs1010697480 | 52 | P>L | No | Ensembl | |
| rs745709625 | 52 | P>S | No |
ExAC gnomAD |
|
| rs781106752 | 53 | Q>P | No |
ExAC gnomAD |
|
| rs781106752 | 53 | Q>R | No |
ExAC gnomAD |
|
| rs757006459 | 54 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1192003856 | 56 | T>I | No |
TOPMed gnomAD |
|
| rs372275031 | 57 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs757988510 | 58 | L>M | No |
ExAC gnomAD |
|
| rs1461484581 | 59 | Q>E | No |
TOPMed gnomAD |
|
| rs752131996 | 59 | Q>P | No |
ExAC gnomAD |
|
| rs766955909 | 60 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs773595959 | 62 | Q>K | No |
ExAC gnomAD |
|
| TCGA novel | 63 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767821305 | 64 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs201004105 | 67 | P>L | No |
1000Genomes ExAC gnomAD |
|
| rs774490241 | 68 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| COSM3529283 | 68 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1599513620 | 69 | G>D | No | Ensembl | |
| COSM709817 | 69 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775411453 | 75 | V>L | No |
ExAC gnomAD |
|
| rs775411453 | 75 | V>M | No |
ExAC gnomAD |
|
| rs769812987 | 76 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1163123624 | 77 | E>D | No | gnomAD | |
| rs757063644 | 79 | P>L | No |
ExAC gnomAD |
|
| rs896958855 | 79 | P>S | No | Ensembl | |
| rs1246982032 | 82 | P>A | No |
TOPMed gnomAD |
|
| rs1246982032 | 82 | P>S | No |
TOPMed gnomAD |
|
|
rs199739375 COSM1287598 |
83 | A>T | autonomic_ganglia [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1005082258 | 84 | P>L | No | TOPMed | |
| rs1202859646 | 84 | P>T | No | gnomAD | |
| rs764610764 | 85 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1267037264 | 88 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs762232183 | 89 | G>D | No |
ExAC gnomAD |
|
| rs943775317 | 89 | G>S | No | TOPMed | |
| rs764426620 | 94 | S>L | No |
ExAC gnomAD |
|
| rs1322966351 | 95 | P>S | No | gnomAD | |
| rs775672646 | 99 | Q>E | No |
ExAC gnomAD |
|
| rs1294507727 | 99 | Q>H | No | gnomAD | |
| rs201999331 | 99 | Q>R | No |
ExAC gnomAD |
|
| rs1568479918 | 100 | Y>C | No | Ensembl | |
| rs759562858 | 101 | I>L | No |
ExAC gnomAD |
|
| rs759562858 | 101 | I>V | No |
ExAC gnomAD |
|
| rs1389177933 | 102 | V>M | No | gnomAD | |
| rs1450930844 | 106 | S>F | No | gnomAD | |
| rs760648471 | 108 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs773081521 | 108 | G>V | No |
ExAC gnomAD |
|
| rs771705262 | 109 | A>T | No |
ExAC gnomAD |
|
| rs747840594 | 109 | A>V | No |
ExAC gnomAD |
|
| rs1018933450 | 110 | M>T | No | Ensembl | |
| rs748847129 | 111 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs757794827 | 111 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs757794827 | 111 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs748847129 | 111 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs778216455 | 114 | E>K | No |
ExAC gnomAD |
|
| rs778216455 | 114 | E>Q | No |
ExAC gnomAD |
|
| COSM4074737 | 115 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs367668632 | 116 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs367668632 | 116 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
|
rs753088781 COSM1680732 |
117 | S>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs765601746 | 119 | A>P | No |
ExAC gnomAD |
|
| rs759798572 | 120 | S>G | No |
ExAC gnomAD |
|
| rs753999610 | 121 | P>L | No |
ExAC gnomAD |
|
| rs753999610 | 121 | P>R | No |
ExAC gnomAD |
|
| rs1296117209 | 121 | P>S | No | TOPMed | |
| rs374515469 | 122 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs371029165 | 122 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs79751004 | 124 | T>P | No | Ensembl | |
| rs141643042 | 125 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1412887190 | 125 | A>V | No | gnomAD | |
| rs768339132 | 129 | G>S | No |
ExAC gnomAD |
|
| rs377552211 | 130 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs377552211 | 130 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1463547768 | 131 | P>L | No |
TOPMed gnomAD |
|
| rs1249964454 | 136 | Q>R | No | gnomAD | |
| rs1451505340 | 137 | Q>* | No | gnomAD | |
| rs1288226371 | 137 | Q>R | No |
TOPMed gnomAD |
|
| rs1230337205 | 139 | Q>* | No | gnomAD | |
| rs747558970 | 139 | Q>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 140 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778213929 | 140 | G>V | No |
ExAC gnomAD |
|
| rs758911880 | 141 | T>A | No |
ExAC gnomAD |
|
| rs144348762 | 141 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1431447142 | 142 | Q>* | No | gnomAD | |
| rs779377898 | 143 | Q>E | No |
ExAC gnomAD |
|
| rs758297278 | 144 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1164488002 | 145 | L>Q | No | gnomAD | |
| rs1382963359 | 147 | V>A | No | gnomAD | |
| rs1397945726 | 149 | T>R | No | TOPMed | |
| rs1184903999 | 150 | S>N | No |
TOPMed gnomAD |
|
| rs765030323 | 150 | S>R | No |
ExAC gnomAD |
|
| rs369384498 | 151 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1341351297 | 153 | A>T | No | TOPMed | |
| rs1247221447 | 154 | K>Q | No | TOPMed | |
| rs760319316 | 154 | K>R | No |
ExAC gnomAD |
|
| rs935801419 | 156 | G>A | No | TOPMed | |
| rs935801419 | 156 | G>D | No | TOPMed | |
| rs149634688 | 158 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs781253828 | 159 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs775742276 | 160 | P>S | No |
ExAC gnomAD |
|
| rs1386249304 | 162 | Q>* | No | gnomAD | |
| rs867580510 | 165 | N>D | No | gnomAD | |
| TCGA novel | 165 | N>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1389060071 | 166 | I>M | No |
TOPMed gnomAD |
|
| rs1444857820 | 166 | I>V | No | gnomAD | |
| rs1190373774 | 167 | Q>* | No | gnomAD | |
| rs1184938428 | 167 | Q>H | No | TOPMed | |
| rs1430264647 | 168 | V>E | No | gnomAD | |
| rs1262297690 | 169 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1209028686 | 170 | Q>P | No | gnomAD | |
| rs1460355861 | 175 | T>M | No | gnomAD | |
|
COSM3822092 rs754841032 |
177 | R>C | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM252777 rs116768203 |
177 | R>H | ovary [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs116768203 | 177 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1568470780 | 179 | V>L | No | Ensembl | |
| rs779664467 | 180 | V>A | No |
ExAC gnomAD |
|
| rs779664467 | 180 | V>G | No |
ExAC gnomAD |
|
| rs750027612 | 181 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs61735284 | 183 | A>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs61735284 RCV000967504 |
183 | A>T | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1443764430 | 184 | A>G | No | TOPMed | |
| rs1599496590 | 186 | G>A | No | Ensembl | |
| rs1388060716 | 186 | G>S | No | Ensembl | |
| rs978609150 | 188 | K>N | No | TOPMed | |
| rs1031720688 | 189 | G>D | No |
TOPMed gnomAD |
|
| rs369848990 | 190 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1326324048 | 190 | G>S | No | gnomAD | |
| rs777089850 | 191 | Q>R | No |
ExAC gnomAD |
|
| rs768610569 | 195 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs748300798 | 197 | H>R | No |
ExAC gnomAD |
|
| rs774273378 | 198 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1335531428 | 198 | G>S | No | gnomAD | |
| rs768660973 | 202 | V>M | No | ExAC | |
| rs201210853 | 204 | S>L | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 204 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1355238878 | 206 | P>A | No | TOPMed | |
| TCGA novel | 206 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs867833290 | 207 | E>K | No | Ensembl | |
| rs769847268 | 209 | S>L | No |
ExAC TOPMed gnomAD |
|
|
COSM1390887 rs1185982655 |
210 | P>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1185982655 | 210 | P>Q | No | gnomAD | |
| rs781108974 | 211 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1452105590 | 211 | V>L | No |
TOPMed gnomAD |
|
| rs770634668 | 212 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs746580616 | 220 | T>R | No |
ExAC gnomAD |
|
| rs757907868 | 222 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1599494572 | 223 | A>P | No | Ensembl | |
| rs1291895484 | 224 | P>S | No | gnomAD | |
| rs752202138 | 225 | T>M | No |
ExAC gnomAD |
|
| rs752202138 | 225 | T>R | No |
ExAC gnomAD |
|
| rs1271803919 | 226 | G>S | No | gnomAD | |
| rs1301030831 | 230 | Q>K | No | gnomAD | |
| rs1447181673 | 230 | Q>R | No | TOPMed | |
| rs1426846404 | 234 | V>A | No | gnomAD | |
|
rs374974524 COSM1390886 |
236 | G>S | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs762098757 | 237 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs762098757 | 237 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1568469668 | 241 | V>I | No | Ensembl | |
| rs143147749 | 243 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs867693847 | 245 | Q>K | No | Ensembl | |
| rs1203228590 | 246 | E>D | No | gnomAD | |
| rs775677549 | 246 | E>K | No |
ExAC gnomAD |
|
| rs1451918382 | 247 | R>K | No |
TOPMed gnomAD |
|
| rs759562849 | 248 | S>P | No |
ExAC gnomAD |
|
| rs866148309 | 249 | V>A | No | Ensembl | |
| rs912782986 | 250 | V>I | No | Ensembl | |
| TCGA novel | 251 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1470587278 | 252 | A>T | No |
TOPMed gnomAD |
|
| rs1363627218 | 252 | A>V | No | gnomAD | |
| rs1432249521 | 256 | A>T | No | gnomAD | |
| rs776771970 | 256 | A>V | No |
ExAC gnomAD |
|
| TCGA novel | 258 | K>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1022161716 | 258 | K>N | No | Ensembl | |
| rs1416967577 | 259 | P>H | No | gnomAD | |
| rs1416967577 | 259 | P>R | No | gnomAD | |
| rs760430294 | 260 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1443540268 | 261 | P>A | No | gnomAD | |
| rs199915135 | 261 | P>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199915135 | 261 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1469541067 | 263 | Q>P | No | gnomAD | |
| rs1483521552 | 264 | P>A | No |
TOPMed gnomAD |
|
| rs1555752506 | 264 | P>L | No | Ensembl | |
| rs1555752506 | 264 | P>R | No | Ensembl | |
| rs1483521552 | 264 | P>S | No |
TOPMed gnomAD |
|
| rs771647665 | 265 | L>P | No |
ExAC gnomAD |
|
| rs748781641 | 269 | G>A | No |
ExAC gnomAD |
|
| rs537753201 | 269 | G>R | No |
1000Genomes ExAC gnomAD |
|
| rs537753201 | 269 | G>S | No |
1000Genomes ExAC gnomAD |
|
| rs1317211829 | 270 | L>F | No | gnomAD | |
| rs199880525 | 270 | L>P | No |
ExAC gnomAD |
|
| rs757749092 | 272 | P>T | No |
ExAC gnomAD |
|
| rs1346142322 | 273 | V>A | No | gnomAD | |
| rs747379983 | 273 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs149107931 | 274 | H>R | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 276 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1184852674 | 277 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs761661391 | 280 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs751322570 | 281 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1165630226 | 282 | L>F | No | gnomAD | |
| rs920692481 | 283 | Q>P | No | Ensembl | |
| rs376946651 | 284 | Q>H | No |
ESP TOPMed gnomAD |
|
| rs1424207005 | 284 | Q>P | No | gnomAD | |
| rs763782834 | 285 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs763782834 | 285 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1472612997 | 286 | P>R | No | TOPMed | |
| rs972067969 | 286 | P>S | No | Ensembl | |
| rs774889704 | 287 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs758960034 | 291 | Y>F | No |
ExAC gnomAD |
|
| rs1248533996 | 291 | Y>N | No | gnomAD | |
|
rs951951549 COSM1494228 |
293 | S>R | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated TOPMed NCI-TCGA Cosmic |
| rs1222207093 | 294 | Q>R | No | gnomAD | |
| rs772677710 | 295 | V>L | No |
ExAC gnomAD |
|
| rs769032243 | 296 | Q>P | No |
ExAC gnomAD |
|
| rs769032243 | 296 | Q>R | No |
ExAC gnomAD |
|
| rs775587157 | 298 | V>A | No | Ensembl | |
| TCGA novel | 299 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749461046 | 300 | G>S | No |
ExAC gnomAD |
|
| rs1448269712 | 301 | G>S | No | gnomAD | |
| rs1344996957 | 302 | D>A | No | TOPMed | |
| rs1336070336 | 302 | D>N | No |
TOPMed gnomAD |
|
| rs773817563 | 303 | A>T | No | Ensembl | |
| rs781145487 | 306 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1362209739 | 308 | S>I | No | gnomAD | |
| rs374107792 | 309 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs770763508 | 310 | I>T | No |
TOPMed gnomAD |
|
|
COSM4426696 rs1034202672 |
311 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs371257748 | 311 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1002291699 | 312 | S>C | No |
TOPMed gnomAD |
|
| rs1429932350 | 313 | S>N | No | gnomAD | |
| rs1324506709 | 315 | Y>C | No | gnomAD | |
| rs1324506709 | 315 | Y>F | No | gnomAD | |
| rs1324506709 | 315 | Y>S | No | gnomAD | |
| rs1299787267 | 318 | P>S | No | gnomAD | |
| rs780486654 | 319 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs764991643 | 320 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1599484867 | 320 | T>P | No | Ensembl | |
| rs765922976 | 321 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs892403741 | 321 | P>S | No |
TOPMed gnomAD |
|
| rs1464192936 | 323 | Y>H | No | TOPMed | |
| rs1599484803 | 323 | Y>S | No | Ensembl | |
| rs775057554 | 324 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1599484795 | 324 | T>P | No | Ensembl | |
| rs367733057 | 325 | Q>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs928961990 | 326 | T>A | No | Ensembl | |
|
COSM6083688 COSM565129 rs775693470 |
326 | T>M | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs928961990 | 326 | T>P | No | Ensembl | |
| rs775693470 | 326 | T>R | No |
ExAC gnomAD |
|
| rs746046482 | 327 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1599484747 | 328 | S>T | No | Ensembl | |
| rs1291851466 | 329 | T>A | No | gnomAD | |
| rs770937959 | 329 | T>I | No | ExAC | |
| rs1291851466 | 329 | T>P | No | gnomAD | |
| rs1037325815 | 330 | S>N | No |
TOPMed gnomAD |
|
| rs747074510 | 330 | S>R | No |
ExAC gnomAD |
|
| rs758249269 | 332 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs567442354 | 333 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs753483809 | 335 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs753483809 | 335 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs986044699 | 335 | A>V | No |
TOPMed gnomAD |
|
| rs766118274 | 337 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs749920960 | 340 | Q>R | No |
ExAC gnomAD |
|
| rs925374458 | 341 | V>A | No | TOPMed | |
| rs763319107 | 341 | V>I | No |
ExAC gnomAD |
|
| rs753133767 | 343 | T>N | No |
ExAC gnomAD |
|
| rs534259784 | 343 | T>P | No | 1000Genomes | |
| rs759764744 | 344 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs759764744 | 344 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs373805947 | 345 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1259848409 | 345 | A>V | No | gnomAD | |
| rs760796567 | 346 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs760796567 | 346 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs143238251 | 349 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1568464137 | 349 | A>S | No | Ensembl | |
| rs143238251 | 349 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs768633232 | 351 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1599484538 | 353 | S>G | No | Ensembl | |
| rs1279035855 | 354 | G>C | No | gnomAD | |
| TCGA novel | 354 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1279035855 | 354 | G>S | No | gnomAD | |
| rs1402528669 | 355 | S>F | No | gnomAD | |
| rs755805103 | 356 | M>I | No | ExAC | |
| rs1469710895 | 357 | P>S | No |
TOPMed gnomAD |
|
| rs749976126 | 358 | M>I | No |
ExAC gnomAD |
|
| rs1178625765 | 358 | M>K | No | gnomAD | |
| rs1404422569 | 358 | M>V | No | gnomAD | |
| COSM474227 | 360 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758945274 | 360 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs759956863 | 362 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1214838698 | 363 | S>I | No | TOPMed | |
| TCGA novel | 363 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs551233553 | 366 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201046489 | 367 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs201046489 | 367 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1283892650 | 367 | A>V | No | gnomAD | |
| rs1420452117 | 368 | S>G | No | TOPMed | |
|
VAR_059781 rs2305780 |
370 | T>A | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2305780 | 370 | T>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1200911049 | 372 | T>S | No |
TOPMed gnomAD |
|
| rs779943590 | 375 | G>V | No | ExAC | |
| rs778302721 | 376 | A>V | No | Ensembl | |
| rs769600950 | 379 | S>G | No |
ExAC gnomAD |
|
| rs1381682916 | 381 | G>E | No |
TOPMed gnomAD |
|
| rs201064266 | 381 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751073401 | 382 | G>A | No |
ExAC gnomAD |
|
| rs751073401 | 382 | G>D | No |
ExAC gnomAD |
|
| rs756772282 | 382 | G>S | No |
ExAC gnomAD |
|
| rs755432116 | 384 | G>S | No |
ExAC gnomAD |
|
| rs1599484218 | 386 | G>C | No | Ensembl | |
| rs750668921 | 389 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs750668921 | 389 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs761840229 | 390 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs774466383 | 391 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1287465181 | 392 | G>A | No | gnomAD | |
| rs201914058 | 392 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201914058 | 392 | G>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1397778721 | 393 | G>R | No | gnomAD | |
| rs769651791 | 394 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1362023530 | 394 | G>S | No | gnomAD | |
| rs769651791 | 394 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1166877415 | 395 | G>D | No | gnomAD | |
| rs776211108 | 395 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs776211108 | 395 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs770583538 | 396 | G>C | No |
ExAC gnomAD |
|
| TCGA novel | 396 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746527383 | 397 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs755485281 | 398 | G>A | No |
ExAC gnomAD |
|
| rs755485281 | 398 | G>D | No |
ExAC gnomAD |
|
| rs908706789 | 398 | G>S | No | TOPMed | |
| rs1343667760 | 399 | G>D | No | TOPMed | |
| rs749740958 | 399 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1222010046 | 400 | S>G | No | gnomAD | |
| rs1202673062 | 400 | S>N | No | TOPMed | |
| rs1485764075 | 402 | S>G | No | TOPMed | |
| rs756502909 | 402 | S>R | No |
ExAC gnomAD |
|
| rs767683838 | 403 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs767683838 | 403 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1212008358 | 403 | T>S | No | TOPMed | |
| rs751620307 | 404 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs764203031 | 405 | G>V | No |
ExAC gnomAD |
|
| rs370992224 | 406 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs960634966 | 407 | G>S | No |
TOPMed gnomAD |
|
| rs1403916440 | 408 | S>R | No | gnomAD | |
| rs1388010039 | 408 | S>T | No |
TOPMed gnomAD |
|
| rs199688398 | 409 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs770523702 | 411 | G>D | No |
ExAC gnomAD |
|
| rs374833159 | 414 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1199023211 | 416 | Q>* | No | gnomAD | |
| rs1599483828 | 417 | G>R | No | Ensembl | |
| rs1297359373 | 418 | G>D | No |
TOPMed gnomAD |
|
| rs897581651 | 418 | G>S | No |
TOPMed gnomAD |
|
| rs201143416 | 420 | M>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201143416 | 420 | M>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 420 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780581056 | 421 | L>P | No |
ExAC gnomAD |
|
| rs756623062 | 422 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs756623062 | 422 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs199969477 | 423 | S>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1302770038 | 424 | A>V | No | gnomAD | |
| rs757425139 | 425 | S>G | No |
ExAC gnomAD |
|
| rs1398904642 | 427 | S>F | No | gnomAD | |
| rs751773825 | 428 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs1400830671 | 429 | S>F | No | gnomAD | |
| rs778124562 | 429 | S>P | No |
ExAC gnomAD |
|
| rs1463450475 | 432 | T>I | No | TOPMed | |
| rs77079245 | 433 | R>H | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 434 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1199831583 | 434 | A>V | No | gnomAD | |
| rs1186789874 | 435 | S>L | No | TOPMed | |
| rs1236396971 | 438 | T>M | No | gnomAD | |
| rs753876589 | 440 | Q>K | No |
ExAC gnomAD |
|
| rs766223041 | 440 | Q>R | No |
ExAC gnomAD |
|
| rs1445685204 | 443 | L>V | No | gnomAD | |
| rs1420854756 | 444 | D>H | No | TOPMed | |
| rs1399661079 | 448 | T>M | No |
TOPMed gnomAD |
|
| TCGA novel | 449 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1413161203 | 452 | V>L | No | gnomAD | |
|
COSM1523817 COSM6149491 rs1413161203 |
452 | V>M | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1423419677 | 454 | L>V | No | TOPMed | |
| rs1599480497 | 457 | S>G | No | Ensembl | |
| rs762357137 | 457 | S>N | No |
ExAC gnomAD |
|
| rs762357137 | 457 | S>T | No |
ExAC gnomAD |
|
| rs1369434908 | 458 | T>I | No | gnomAD | |
| rs1599480480 | 458 | T>P | No | Ensembl | |
| rs1599480446 | 460 | Y>S | No | Ensembl | |
| rs1260522590 | 462 | H>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1599480409 | 466 | H>P | No | Ensembl | |
| rs1403202230 | 468 | Q>R | No | TOPMed | |
| rs1446665042 | 470 | Q>H | No | TOPMed | |
| rs747423569 | 474 | P>S | No |
ExAC gnomAD |
|
| rs772430581 | 475 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1599480367 | 476 | N>T | No | Ensembl | |
| TCGA novel | 478 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 479 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs867992954 | 481 | G>D | No | Ensembl | |
| TCGA novel | 482 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 486 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs750251703 COSM1235940 |
487 | V>I | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| COSM3529281 | 488 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767230838 | 489 | M>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 490 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 491 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756915561 | 494 | R>C | No |
ExAC gnomAD |
|
| rs751282140 | 495 | R>H | No |
ExAC gnomAD |
|
| rs868625390 | 510 | R>C | No |
TOPMed gnomAD |
|
| rs751276498 | 510 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1368455521 | 513 | A>P | No | gnomAD | |
| rs1368983698 | 516 | P>A | No | TOPMed | |
| rs1473771376 | 516 | P>L | No | TOPMed | |
| rs952913844 | 517 | L>V | No |
TOPMed gnomAD |
|
| rs1445253062 | 518 | L>Q | No | gnomAD | |
| rs763853027 | 519 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs763853027 | 519 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1379670900 | 519 | R>W | No | gnomAD | |
| TCGA novel | 520 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs866154497 | 521 | M>I | No | gnomAD | |
| rs1357779458 | 521 | M>L | No | gnomAD | |
| rs776268537 | 522 | E>D | No | gnomAD | |
| rs868663484 | 522 | E>G | No | Ensembl | |
| rs1395184240 | 524 | Q>R | No | gnomAD | |
| rs757936541 | 525 | Q>H | No |
ExAC gnomAD |
|
| rs866762330 | 525 | Q>K | No | Ensembl | |
| rs965255423 | 527 | M>I | No | Ensembl | |
| rs866164355 | 527 | M>T | No | Ensembl | |
| rs1177700853 | 529 | M>I | No | gnomAD | |
| rs377047346 | 529 | M>L | No | Ensembl | |
| rs1239873615 | 529 | M>T | No | gnomAD | |
| rs866758516 | 530 | R>L | No | gnomAD | |
| rs866758516 | 530 | R>Q | No | gnomAD | |
| rs752281324 | 530 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1348793042 | 531 | G>C | No | gnomAD | |
| rs1285442837 | 531 | G>D | No | gnomAD | |
| rs759107756 | 533 | P>A | No |
ExAC gnomAD |
|
| rs531624120 | 533 | P>L | No |
1000Genomes ExAC gnomAD |
|
| rs1395953234 | 535 | S>L | No | gnomAD | |
| rs1232507732 | 535 | S>P | No | gnomAD | |
| rs1006678164 | 536 | Q>R | No | Ensembl | |
| COSM3529280 | 544 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754632913 | 544 | Q>P | No |
ExAC gnomAD |
|
| rs754632913 | 544 | Q>R | No |
ExAC gnomAD |
|
| rs139000839 | 545 | K>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs984696567 | 546 | M>I | No | gnomAD | |
| rs1487803502 | 550 | T>I | No | gnomAD | |
| rs1276680000 | 552 | G>D | No | gnomAD | |
| rs1317762657 | 552 | G>S | No |
TOPMed gnomAD |
|
| rs146416788 | 553 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1241659 rs146416788 |
553 | V>M | oesophagus [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
|
rs764655187 COSM4074735 |
554 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs753034014 | 555 | V>G | No |
ExAC gnomAD |
|
| COSM3403826 | 556 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759713486 | 559 | P>L | No |
ExAC TOPMed gnomAD |
|
|
rs148533868 RCV000884900 |
559 | P>S | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1168189308 | 561 | T>A | No | gnomAD | |
| rs543467989 | 561 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs543467989 | 561 | T>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs140854938 | 564 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1054762 | 566 | I>M | No |
ExAC gnomAD |
|
| rs1256600048 | 567 | S>G | No | gnomAD | |
| rs1568460306 | 567 | S>N | No | Ensembl | |
| rs755643451 | 567 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs957621921 | 568 | A>T | No |
TOPMed gnomAD |
|
| rs1599478106 | 570 | V>G | No | Ensembl | |
| TCGA novel | 571 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 571 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376758339 | 572 | Q>L | No |
ESP ExAC gnomAD |
|
| rs1359057551 | 573 | Y>N | No | TOPMed | |
| TCGA novel | 577 | L>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774491041 | 581 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs764088725 | 581 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs774491041 | 581 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1487712980 | 582 | S>G | No | gnomAD | |
| TCGA novel | 583 | L>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs368306820 | 584 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1206834006 | 585 | D>N | No |
TOPMed gnomAD |
|
| rs775302421 | 587 | T>K | No |
ExAC gnomAD |
|
| rs976177265 | 588 | E>K | No | TOPMed | |
| rs151266260 | 590 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs770484234 | 591 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs746492553 | 594 | K>R | No |
ExAC gnomAD |
|
| rs779445963 | 597 | P>L | No |
ExAC gnomAD |
|
| COSM4074734 | 598 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372765441 | 599 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
|
rs550390197 COSM1264055 |
600 | V>I | oesophagus [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs757451059 | 601 | G>A | No |
ExAC gnomAD |
|
| rs146658544 | 601 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs751663921 | 602 | P>T | No |
ExAC gnomAD |
|
| rs199653217 | 603 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM6149492 | 604 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1599475428 COSM1235939 |
605 | I>M | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated Ensembl |
| rs1336751489 | 605 | I>S | No | gnomAD | |
| rs1205967623 | 607 | A>D | No | TOPMed | |
| rs368076467 | 607 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs776409553 | 608 | F>L | No |
ExAC gnomAD |
|
| rs770551512 | 609 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs769159405 | 613 | R>Q | No |
ExAC gnomAD |
|
| rs1254163149 | 615 | H>D | No | TOPMed | |
| COSM6083689 | 617 | E>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749748603 | 617 | E>D | No |
ExAC gnomAD |
|
| rs1276622350 | 618 | A>P | No | gnomAD | |
| rs778126997 | 619 | I>V | No |
ExAC gnomAD |
|
| COSM4721203 | 621 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 621 | D>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746973928 | 622 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs766107840 | 623 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs143203128 | 623 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs753634327 | 623 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs143203128 | 623 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1599475094 | 624 | V>G | No | Ensembl | |
| rs755894311 | 624 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1420043564 | 627 | Q>* | No | TOPMed | |
| rs532429887 | 633 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs773682844 | 636 | K>Q | No |
ExAC gnomAD |
|
|
rs930407483 COSM3970736 |
641 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1599475012 | 641 | Y>D | No | Ensembl | |
| rs148634929 | 642 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs144971619 | 643 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs144971619 | 643 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs956897703 | 646 | P>S | No | Ensembl | |
| rs1396060414 | 648 | E>K | No | TOPMed | |
| rs747372190 | 649 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1228161235 | 651 | P>L | No | gnomAD | |
| rs1213323538 | 654 | V>I | No | gnomAD | |
| rs369726740 | 657 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs369726740 | 657 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs267605300 | 658 | A>T | No | Ensembl | |
| TCGA novel | 659 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376479334 | 659 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 661 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs942094601 | 661 | R>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 663 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1327814768 | 664 | K>Q | No | gnomAD | |
| rs199887401 | 669 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs199887401 | 669 | L>R | No |
ExAC TOPMed gnomAD |
|
| COSM1480660 | 670 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767829998 | 674 | E>D | No |
ExAC gnomAD |
|
| rs1175398818 | 674 | E>K | No | gnomAD | |
| rs1175398818 | 674 | E>Q | No | gnomAD | |
|
COSM991785 rs371857825 |
676 | V>M | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1457650605 | 677 | L>I | No | gnomAD | |
| rs775504584 | 680 | T>S | No |
ExAC gnomAD |
|
| rs1227352957 | 682 | H>R | No | TOPMed | |
| rs201598179 | 686 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1232992870 | 688 | Y>* | No | gnomAD | |
| rs1599473724 | 688 | Y>H | No | Ensembl | |
| rs781172033 | 690 | G>S | No |
ExAC gnomAD |
|
| rs770697492 | 690 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1334961887 | 692 | V>L | No | gnomAD | |
| COSM1390885 | 694 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1276509692 | 697 | P>S | No | TOPMed | |
| rs777338298 | 698 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs747668425 | 699 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs747668425 | 699 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs778511023 | 701 | R>Q | No |
ExAC gnomAD |
|
| COSM991784 | 701 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 702 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 705 | S>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1180352144 | 705 | S>N | No | gnomAD | |
| rs886525443 | 707 | L>F | No |
TOPMed gnomAD |
|
| rs760603316 | 708 | T>N | No |
ExAC gnomAD |
|
| rs1476404146 | 709 | Q>H | No | gnomAD | |
| rs1430154551 | 710 | A>V | No |
TOPMed gnomAD |
|
| rs1327175076 | 712 | R>W | No | TOPMed | |
| COSM438703 | 715 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1320921344 | 716 | K>R | No | gnomAD | |
| rs1242806184 | 718 | L>Q | No | gnomAD | |
| rs747921796 | 719 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1039685384 | 724 | H>R | No |
TOPMed gnomAD |
|
| rs1568457833 | 725 | A>T | No | Ensembl | |
| rs779546732 | 726 | M>L | No |
ExAC gnomAD |
|
| rs779546732 | 726 | M>V | No |
ExAC gnomAD |
|
| rs771351477 | 727 | V>I | No |
ExAC gnomAD |
|
| rs1568457810 | 728 | N>S | No | Ensembl | |
| rs1447474224 | 731 | E>A | No | gnomAD | |
| rs563474870 | 731 | E>D | No |
1000Genomes ExAC gnomAD |
|
| rs778245794 | 734 | L>V | No |
ExAC gnomAD |
|
| rs758801717 | 735 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs188957964 RCV000959272 |
735 | R>Q | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs758801717 | 735 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1337547352 | 736 | V>G | No | gnomAD | |
|
rs377533889 COSM6149493 COSM1523819 |
736 | V>L | lung large_intestine Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated ESP ExAC TOPMed gnomAD NCI-TCGA Cosmic |
| rs377533889 | 736 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs933760047 | 738 | V>G | No | TOPMed | |
| rs1005366518 | 738 | V>M | No | Ensembl | |
| rs376569607 | 739 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs372703326 | 740 | A>V | No |
ESP TOPMed gnomAD |
|
| rs761835722 | 743 | A>G | No | Ensembl | |
| rs1349177606 | 743 | A>T | No | gnomAD | |
| rs780110843 | 744 | F>L | No | ExAC | |
| rs756267517 | 745 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 746 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs972232903 | 747 | T>I | No | TOPMed | |
| rs1409995542 | 750 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs909491359 | 752 | T>A | No | TOPMed | |
| rs1310502794 | 752 | T>R | No | gnomAD | |
| rs1359720795 | 754 | L>F | No | gnomAD | |
| rs985059962 | 755 | N>K | No |
TOPMed gnomAD |
|
| rs1183179159 | 758 | A>V | No | gnomAD | |
| TCGA novel | 760 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1190214573 | 761 | A>S | No | gnomAD | |
|
COSM3822091 rs1483602267 |
761 | A>V | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1407974879 | 762 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs768449350 | 769 | A>E | No | gnomAD | |
| rs1319124512 | 769 | A>T | No | gnomAD | |
| rs768449350 | 769 | A>V | No | gnomAD | |
| rs1423614688 | 772 | N>S | No | TOPMed | |
| rs751409797 | 773 | Q>R | No |
ExAC gnomAD |
|
| rs910823699 | 776 | S>N | No | Ensembl | |
| rs1002857882 | 776 | S>R | No | TOPMed | |
| rs1379253084 | 779 | N>K | No | gnomAD | |
| TCGA novel | 781 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs372349612 | 783 | F>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs775244921 | 785 | N>S | No |
ExAC gnomAD |
|
| rs1167365363 | 786 | V>M | No | gnomAD | |
| rs1199963737 | 789 | Q>E | No |
TOPMed gnomAD |
|
| rs927523898 | 791 | S>* | No |
TOPMed gnomAD |
|
| rs927523898 | 791 | S>L | No |
TOPMed gnomAD |
|
| rs1175106112 | 794 | C>G | No | gnomAD | |
| rs749697367 | 795 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1274646373 | 795 | R>S | No | gnomAD | |
| rs1257671602 | 797 | E>* | No |
TOPMed gnomAD |
|
| rs1257671602 | 797 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 798 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1225476314 | 798 | D>N | No | TOPMed | |
| rs559198056 | 799 | R>P | No |
1000Genomes ExAC gnomAD |
|
| rs867862091 | 799 | R>S | No | Ensembl | |
| rs865905618 | 800 | V>L | No | Ensembl | |
| rs1005332241 | 802 | Q>R | No | Ensembl | |
| rs781556786 | 805 | E>G | No |
ExAC gnomAD |
|
| TCGA novel | 807 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201362842 | 810 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201362842 | 810 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs747034757 | 811 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs747034757 | 811 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs777787788 | 814 | Q>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 817 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1473347782 | 818 | L>Q | No | gnomAD | |
| rs1251940908 | 822 | A>T | No | gnomAD | |
| rs752540965 | 823 | A>S | No |
ExAC gnomAD |
|
| rs993851745 | 823 | A>V | No | Ensembl | |
| rs147242039 | 827 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147242039 | 827 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1382498013 | 828 | V>L | No | TOPMed | |
| rs753529527 | 831 | Q>R | No |
ExAC gnomAD |
|
| rs1292879865 | 833 | L>F | No | gnomAD | |
| rs1292879865 | 833 | L>V | No | gnomAD | |
| rs764669877 | 837 | Q>R | No |
ExAC gnomAD |
|
| rs776056625 | 838 | G>D | No |
ExAC gnomAD |
|
| rs369260631 | 839 | S>G | No | Ensembl | |
| rs759945290 | 840 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs759945290 | 840 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1311519725 | 841 | G>D | No | gnomAD | |
| rs747142189 | 841 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs747142189 | 841 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1599470097 | 850 | L>F | No | Ensembl | |
| rs1465154240 | 851 | L>F | No | gnomAD | |
| rs753582886 | 852 | K>E | No |
ExAC gnomAD |
|
| rs1204953321 | 856 | Y>C | No | TOPMed | |
| TCGA novel | 858 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1278785640 | 859 | M>T | No | gnomAD | |
| rs754267216 | 862 | R>W | No |
ExAC gnomAD |
|
| rs1275131556 | 866 | L>P | No | gnomAD | |
| TCGA novel | 866 | L>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM991780 | 867 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 867 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1348123513 | 868 | S>N | No | gnomAD | |
| rs1409879987 | 870 | A>G | No | gnomAD | |
| rs376362982 | 878 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1425540118 | 878 | I>T | No | gnomAD | |
| rs1317248124 | 879 | R>Q | No | gnomAD | |
| TCGA novel | 879 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1485367521 | 882 | Y>C | No | gnomAD | |
| COSM5516827 | 883 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3281470 | 883 | D>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1343489849 | 884 | E>G | No | TOPMed | |
| rs769521923 | 890 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1313164695 | 890 | I>S | No | gnomAD | |
| COSM4820876 | 891 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1222876052 | 892 | H>D | No | gnomAD | |
| rs571560628 | 893 | R>H | No | 1000Genomes | |
| rs373669117 | 894 | V>I | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 895 | A>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1354259908 | 895 | A>T | No | gnomAD | |
| rs756891688 | 896 | Q>R | No |
ExAC gnomAD |
|
| rs757967033 | 897 | A>D | No |
ExAC gnomAD |
|
| rs757967033 | 897 | A>V | No |
ExAC gnomAD |
|
| rs754461637 | 898 | K>R | No |
ExAC gnomAD |
|
|
rs766888168 CA054984 RCV000201377 |
899 | G>S | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs750760412 | 900 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs767639250 | 901 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1242025414 | 903 | I>T | No | gnomAD | |
| rs761967952 | 905 | V>A | No |
ExAC gnomAD |
|
| rs1191116434 | 905 | V>I | No | TOPMed | |
| rs764229128 | 907 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1228565641 | 907 | G>D | No | gnomAD | |
| rs764229128 | 907 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs764229128 | 907 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1309905818 | 908 | E>* | No |
TOPMed gnomAD |
|
| COSM6083690 | 908 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1309905818 | 908 | E>K | No |
TOPMed gnomAD |
|
| rs1188229507 | 909 | F>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1192556311 | 910 | A>S | No |
TOPMed gnomAD |
|
| rs1488528508 | 910 | A>V | No | gnomAD | |
| rs953834496 | 911 | N>H | No | Ensembl | |
| rs746451025 | 911 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs757663788 | 917 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1232350807 | 917 | N>S | No | gnomAD | |
| rs1303699934 | 918 | P>L | No |
TOPMed gnomAD |
|
| rs751876803 | 919 | L>M | No |
ExAC gnomAD |
|
| rs778006176 | 919 | L>P | No |
ExAC gnomAD |
|
| rs1371754732 | 921 | P>A | No | gnomAD | |
| rs796123607 | 921 | P>L | No | gnomAD | |
| TCGA novel | 922 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs976529787 | 923 | K>N | No | TOPMed | |
| rs1210759805 | 924 | D>E | No | gnomAD | |
| rs1464907286 | 924 | D>N | No |
TOPMed gnomAD |
|
| rs1464907286 | 924 | D>Y | No |
TOPMed gnomAD |
|
| rs1329936811 | 925 | E>K | No | gnomAD | |
| rs1283633503 | 925 | E>V | No | gnomAD | |
| rs748720997 | 926 | E>D | No |
ExAC gnomAD |
|
| rs1342824790 | 929 | E>* | No | gnomAD | |
| rs1568454994 | 929 | E>D | No | Ensembl | |
| rs1342824790 | 929 | E>K | No | gnomAD | |
| rs1457067172 | 930 | E>K | No | gnomAD | |
| rs1314062056 | 931 | E>K | No | gnomAD | |
| rs1314062056 | 931 | E>Q | No | gnomAD | |
| rs922670319 | 932 | E>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1386839521 | 933 | S>R | No | gnomAD | |
| rs201266626 | 935 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs771391827 | 935 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1385084150 | 937 | L>M | No | gnomAD | |
| rs778057637 | 937 | L>Q | No |
ExAC gnomAD |
|
| rs753031607 | 938 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs201695501 | 938 | P>S | No |
1000Genomes ExAC gnomAD |
|
| rs1197182607 | 939 | Q>K | No | TOPMed | |
| rs984452483 | 940 | D>H | No |
TOPMed gnomAD |
|
| rs984452483 | 940 | D>N | No |
TOPMed gnomAD |
|
| rs1190785377 | 944 | A>S | No | gnomAD | |
| rs755919768 | 944 | A>V | No |
TOPMed gnomAD |
|
| rs1289402865 | 945 | A>D | No | gnomAD | |
| rs1010879284 | 946 | G>A | No |
TOPMed gnomAD |
|
| rs1266562479 | 947 | G>D | No | gnomAD | |
| rs1297886336 | 947 | G>S | No | TOPMed | |
| rs778972180 | 948 | E>Q | No |
ExAC gnomAD |
|
| rs1300214149 | 950 | P>A | No | gnomAD | |
| rs1568454797 | 951 | A>V | No | Ensembl | |
| rs752517709 | 953 | G>D | No |
TOPMed gnomAD |
|
| rs753821927 | 954 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1164135367 | 955 | E>D | No | gnomAD | |
| rs1238958950 | 955 | E>Q | No | TOPMed | |
| rs111774835 | 957 | L>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs111774835 | 957 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1219650054 | 960 | P>R | No | TOPMed | |
| rs970495619 | 961 | A>T | No | TOPMed | |
| rs1266541116 | 964 | A>T | No | TOPMed | |
| rs1246406817 | 967 | D>G | No | gnomAD | |
| rs1475616768 | 967 | D>N | No | gnomAD | |
| rs1024720869 | 968 | A>G | No |
TOPMed gnomAD |
|
| rs1196956794 | 968 | A>S | No |
TOPMed gnomAD |
|
| rs1196956794 | 968 | A>T | No |
TOPMed gnomAD |
|
| rs905953511 | 969 | R>C | No | Ensembl | |
| rs1205107407 | 969 | R>P | No | gnomAD | |
| rs1469617012 | 970 | G>R | No |
TOPMed gnomAD |
|
| TCGA novel | 971 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1048495559 | 972 | F>L | No |
TOPMed gnomAD |
|
| rs1014468530 | 973 | V>G | No | TOPMed | |
| rs1229686716 | 976 | L>M | No |
TOPMed gnomAD |
|
| rs1229686716 | 976 | L>V | No |
TOPMed gnomAD |
No associated diseases with P22670
No regional properties for P22670
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P22670 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR12619 | RFX TRANSCRIPTION FACTOR FAMILY |
| PANTHER Subfamily | PTHR12619:SF23 | MHC CLASS II REGULATORY FACTOR RFX1 |
| PANTHER Protein Class |
DNA-binding transcription factor
helix-turn-helix transcription factor winged helix/forkhead transcription factor |
|
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| immune response | Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P48378 | RFX2 | DNA-binding protein RFX2 | Homo sapiens (Human) | SS |
| P48380 | RFX3 | Transcription factor RFX3 | Homo sapiens (Human) | SS |
| P48381 | Rfx3 | Transcription factor RFX3 | Mus musculus (Mouse) | SS |
| P48377 | Rfx1 | MHC class II regulatory factor RFX1 | Mus musculus (Mouse) | SS |
| Q0V9K5 | rfx3 | Transcription factor RFX3 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MATQAYTELQ | AAPPPSQPPQ | APPQAQPQPP | PPPPPAAPQP | PQPPTAAATP | QPQYVTELQS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PQPQAQPPGG | QKQYVTELPA | VPAPSQPTGA | PTPSPAPQQY | IVVTVSEGAM | RASETVSEAS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PGSTASQTGV | PTQVVQQVQG | TQQRLLVQTS | VQAKPGHVSP | LQLTNIQVPQ | QALPTQRLVV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QSAAPGSKGG | QVSLTVHGTQ | QVHSPPEQSP | VQANSSSSKT | AGAPTGTVPQ | QLQVHGVQQS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VPVTQERSVV | QATPQAPKPG | PVQPLTVQGL | QPVHVAQEVQ | QLQQVPVPHV | YSSQVQYVEG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GDASYTASAI | RSSTYSYPET | PLYTQTASTS | YYEAAGTATQ | VSTPATSQAV | ASSGSMPMYV |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SGSQVVASST | STGAGASNSS | GGGGSGGGGG | GGGGGGGGGS | GSTGGGGSGA | GTYVIQGGYM |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LGSASQSYSH | TTRASPATVQ | WLLDNYETAE | GVSLPRSTLY | CHYLLHCQEQ | KLEPVNAASF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GKLIRSVFMG | LRTRRLGTRG | NSKYHYYGLR | IKASSPLLRL | MEDQQHMAMR | GQPFSQKQRL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| KPIQKMEGMT | NGVAVGQQPS | TGLSDISAQV | QQYQQFLDAS | RSLPDFTELD | LQGKVLPEGV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GPGDIKAFQV | LYREHCEAIV | DVMVNLQFTL | VETLWKTFWR | YNLSQPSEAP | PLAVHDEAEK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| RLPKAILVLL | SKFEPVLQWT | KHCDNVLYQG | LVEILIPDVL | RPIPSALTQA | IRNFAKSLES |
| 730 | 740 | 750 | 760 | 770 | 780 |
| WLTHAMVNIP | EEMLRVKVAA | AGAFAQTLRR | YTSLNHLAQA | ARAVLQNTAQ | INQMLSDLNR |
| 790 | 800 | 810 | 820 | 830 | 840 |
| VDFANVQEQA | SWVCRCEDRV | VQRLEQDFKV | TLQQQNSLEQ | WAAWLDGVVS | QVLKPYQGSA |
| 850 | 860 | 870 | 880 | 890 | 900 |
| GFPKAAKLFL | LKWSFYSSMV | IRDLTLRSAA | SFGSFHLIRL | LYDEYMYYLI | EHRVAQAKGE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| TPIAVMGEFA | NLATSLNPLD | PDKDEEEEEE | EESEDELPQD | ISLAAGGESP | ALGPETLEPP |
| 970 | |||||
| AKLARTDARG | LFVQALPSS |