P21709
Gene name |
EPHA1 (EPH, EPHT, EPHT1) |
Protein name |
Ephrin type-A receptor 1 |
Names |
hEpha1 , EC 2.7.10.1 , EPH tyrosine kinase , EPH tyrosine kinase 1 , Erythropoietin-producing hepatoma receptor , Tyrosine-protein kinase receptor EPH |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2041 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
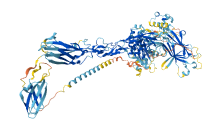
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
624-884 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
613-880 (Protein kinase domain) |
Relief mechanism |
Others |
Assay |
|
Accessory elements
No accessory elements
References
- Hubbard SR (2004) "Juxtamembrane autoinhibition in receptor tyrosine kinases", Nature reviews. Molecular cell biology, 5, 464-71
- Wybenga-Groot LE et al. (2001) "Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region", Cell, 106, 745-57
- Artim SC et al. (2012) "Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family", The Biochemical journal, 448, 213-20
- Uchikawa E et al. (2019) "Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex", eLife, 8,
- Nielsen J et al. (2022) "Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling", Journal of molecular biology, 434, 167458
- Chen YS et al. (2021) "Insertion of a synthetic switch into insulin provides metabolite-dependent regulation of hormone-receptor activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Craddock BP et al. (2007) "Autoinhibition of the insulin-like growth factor I receptor by the juxtamembrane region", FEBS letters, 581, 3235-40
Autoinhibited structure

Activated structure

5 structures for P21709
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2K1K | NMR | - | A/B | 536-573 | PDB |
| 2K1L | NMR | - | A/B | 536-573 | PDB |
| 3HIL | X-ray | 200 A | A/B | 911-974 | PDB |
| 3KKA | X-ray | 240 A | A/B | 911-974 | PDB |
| AF-P21709-F1 | Predicted | AlphaFoldDB |
1164 variants for P21709
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1473690516 | 1 | M>? | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1805636910 | 3 | R>Q | No | TOPMed | |
| rs1805637071 | 3 | R>W | No |
TOPMed gnomAD |
|
| rs1805636800 | 4 | R>L | No | gnomAD | |
| rs1409667882 | 5 | W>* | No | gnomAD | |
| rs1191226865 | 5 | W>G | No |
TOPMed gnomAD |
|
| rs2116647507 | 10 | G>V | No | Ensembl | |
| rs2116647488 | 12 | V>L | No | Ensembl | |
| rs1805635665 | 14 | L>P | No | TOPMed | |
| rs1805635582 | 15 | L>V | No |
TOPMed gnomAD |
|
| rs1343287730 | 17 | A>T | No | TOPMed | |
| rs1805634837 | 18 | P>L | No | Ensembl | |
| rs896909240 | 18 | P>S | No | 1000Genomes | |
| rs1462823087 | 20 | P>H | No | TOPMed | |
| rs1462823087 | 20 | P>L | No | TOPMed | |
| rs1805634512 | 20 | P>S | No | TOPMed | |
| rs1805634253 | 21 | P>A | No | TOPMed | |
| rs1805634253 | 21 | P>S | No | TOPMed | |
| rs1038171832 | 23 | A>P | No |
TOPMed gnomAD |
|
|
RCV000950730 rs79587607 |
24 | R>H | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs79587607 | 24 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2116647348 | 24 | R>S | No | Ensembl | |
| rs1467190935 | 25 | A>P | No |
TOPMed gnomAD |
|
| rs1467190935 | 25 | A>S | No |
TOPMed gnomAD |
|
| rs1273047972 | 26 | K>* | No | TOPMed | |
| rs1273047972 | 26 | K>Q | No | TOPMed | |
| TCGA novel | 29 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs889892936 | 30 | L>P | No | Ensembl | |
| COSM3922987 | 31 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1342428064 | 31 | M>T | No |
TOPMed gnomAD |
|
| rs1050291925 | 32 | D>G | No |
TOPMed gnomAD |
|
| rs774079359 | 34 | S>N | No |
ExAC gnomAD |
|
| rs1000636068 | 35 | K>N | No | TOPMed | |
| rs768501447 | 38 | G>R | No |
ExAC gnomAD |
|
| rs779885990 | 39 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs934062647 | 40 | L>P | No | Ensembl | |
| rs1805591242 | 41 | G>A | No | gnomAD | |
| rs1389840241 | 41 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs755797395 | 45 | D>H | No |
ExAC TOPMed gnomAD |
|
|
rs571770871 COSM452536 COSM452537 |
46 | P>S | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1805590949 | 47 | P>R | No | Ensembl | |
| rs1188031745 | 48 | K>N | No | TOPMed | |
| rs1037157442 | 49 | D>G | No | gnomAD | |
| rs780744262 | 50 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs2116632000 | 53 | E>Q | No | Ensembl | |
| rs1805423738 | 55 | Q>K | No | Ensembl | |
| rs1470179846 | 56 | Q>K | No | gnomAD | |
| rs2116631952 | 57 | I>M | No | Ensembl | |
| rs769627585 | 57 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1224513857 | 60 | G>R | No | gnomAD | |
| rs2116631911 | 61 | T>R | No | Ensembl | |
| rs1805422631 | 61 | T>S | No | Ensembl | |
| rs139238731 | 62 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs139238731 | 62 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs778371838 | 63 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1414693803 | 64 | Y>C | No | Ensembl | |
| rs1319809963 | 65 | M>I | No | gnomAD | |
| rs1805421696 | 65 | M>T | No | Ensembl | |
| rs376995026 | 65 | M>V | No |
ESP gnomAD |
|
| rs758867460 | 67 | Q>* | No |
ExAC gnomAD |
|
| COSM1448852 | 67 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1346434969 | 68 | D>N | No | gnomAD | |
| rs2116631813 | 68 | D>V | No | Ensembl | |
| rs1346434969 | 68 | D>Y | No | gnomAD | |
| TCGA novel | 70 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs118129834 | 71 | M>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1805421137 | 71 | M>T | No | TOPMed | |
| rs2116631774 | 72 | Q>* | No | Ensembl | |
| rs1805420933 | 72 | Q>H | No |
TOPMed gnomAD |
|
| rs2116631764 | 72 | Q>L | No | Ensembl | |
| rs2116631764 | 72 | Q>R | No | Ensembl | |
| rs373319975 | 73 | G>R | No |
ESP TOPMed gnomAD |
|
| rs755258725 | 74 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs755258725 | 74 | R>G | No |
ExAC gnomAD |
|
| rs535622332 | 74 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs535622332 | 74 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1254092990 | 75 | R>K | No | gnomAD | |
| rs766451992 | 75 | R>S | No |
ExAC gnomAD |
|
| rs1563119657 | 77 | T>A | No | Ensembl | |
| rs760843035 | 77 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs760843035 | 77 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs2116631691 | 80 | W>* | No | Ensembl | |
| rs773136368 | 80 | W>* | No |
ExAC TOPMed gnomAD |
|
|
rs74721927 COSM3922983 |
82 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
COSM4762225 rs1029805928 |
82 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1029805928 | 82 | R>L | No | TOPMed | |
| rs1372113896 | 83 | S>T | No | gnomAD | |
| rs1296665931 | 84 | N>D | No |
TOPMed gnomAD |
|
| rs142191815 | 84 | N>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs961514735 | 84 | N>K | No | gnomAD | |
| rs142191815 | 84 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1364023704 | 85 | W>R | No | gnomAD | |
| rs769720234 | 85 | W>S | No |
ExAC gnomAD |
|
| rs1805418104 | 86 | I>L | No | TOPMed | |
| rs1478817861 | 86 | I>M | No |
TOPMed gnomAD |
|
| rs1805418032 | 86 | I>T | No | TOPMed | |
| rs1194676814 | 87 | Y>* | No |
TOPMed gnomAD |
|
| rs2116631573 | 87 | Y>F | No | Ensembl | |
| rs759350123 | 88 | R>C | No |
ExAC gnomAD |
|
| rs148653512 | 88 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs148653512 | 88 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 88 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772621246 | 89 | G>E | No |
ExAC gnomAD |
|
| rs144146635 | 89 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs144146635 | 89 | G>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1805416951 | 90 | E>K | No | TOPMed | |
| rs1486155232 | 91 | E>D | No | Ensembl | |
| rs748648935 | 91 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1805416620 | 92 | A>T | No | TOPMed | |
| rs2116631473 | 92 | A>V | No | Ensembl | |
| COSM5406845 | 93 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2116631453 | 93 | S>P | No | Ensembl | |
|
COSM4587212 rs755348512 |
94 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs755348512 | 94 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs45447297 | 94 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs45447297 | 94 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201228574 | 95 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201228574 | 95 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs752699309 | 97 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs761922296 | 97 | V>L | No |
ExAC gnomAD |
|
| rs761922296 | 97 | V>M | No |
ExAC gnomAD |
|
| rs147254019 | 99 | L>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs143810053 | 100 | Q>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs924106822 | 100 | Q>P | No | Ensembl | |
| rs1805414436 | 102 | T>N | No | TOPMed | |
| rs760227816 | 103 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs760227816 COSM1205425 COSM1205426 |
103 | V>M | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs369887921 | 104 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs369887921 | 104 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs201546539 | 104 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1805413410 | 105 | D>E | No | Ensembl | |
| rs1460792502 | 105 | D>G | No | gnomAD | |
| rs2116631238 | 106 | C>Y | No | Ensembl | |
| rs749711811 | 107 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs780269428 | 107 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1053367957 | 109 | F>S | No | TOPMed | |
| COSM3635281 | 110 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756424890 | 111 | G>E | No |
ExAC TOPMed gnomAD |
|
|
COSM94193 rs1805412828 |
111 | G>R | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
|
COSM362590 rs756424890 COSM362591 |
111 | G>V | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
COSM3635279 rs781509885 |
112 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1475628798 | 113 | A>S | No | gnomAD | |
| rs1475628798 | 113 | A>T | No | gnomAD | |
| rs2116631153 | 113 | A>V | No | Ensembl | |
| rs530283884 | 114 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs759367292 | 115 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1563119463 | 116 | L>P | No | Ensembl | |
| rs972897874 | 117 | G>R | No |
TOPMed gnomAD |
|
| rs972897874 | 117 | G>S | No |
TOPMed gnomAD |
|
| rs1233382933 | 118 | C>Y | No |
TOPMed gnomAD |
|
| rs961153147 | 119 | K>R | No | TOPMed | |
| rs1349425480 | 120 | E>K | No | gnomAD | |
| rs760451814 | 121 | T>N | No |
ExAC gnomAD |
|
| rs772864439 | 122 | F>I | No |
ExAC gnomAD |
|
| rs764543064 | 123 | N>T | No | Ensembl | |
| rs1375957847 | 124 | L>F | No |
TOPMed gnomAD |
|
| rs1299141462 | 125 | L>P | No |
TOPMed gnomAD |
|
| rs1299141462 | 125 | L>Q | No |
TOPMed gnomAD |
|
| rs1392943742 | 126 | Y>C | No |
TOPMed gnomAD |
|
| rs1392943742 | 126 | Y>F | No |
TOPMed gnomAD |
|
|
COSM3635277 rs111858878 |
127 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs767106252 | 127 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs767106252 | 127 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs761342435 | 128 | E>A | No |
ExAC gnomAD |
|
| rs1586585702 | 130 | D>N | No | Ensembl | |
| rs1287337651 | 133 | V>G | No | gnomAD | |
| COSM745160 | 133 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs773830927 | 133 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1158975064 | 135 | I>L | No | Ensembl | |
| rs1805408917 | 137 | L>F | No | Ensembl | |
|
rs1421157458 COSM3411731 COSM3411732 |
138 | R>* | Variant assessed as Somatic; HIGH impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM1448849 COSM1448850 rs190437539 |
138 | R>Q | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| COSM6176849 | 139 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770072024 | 139 | R>P | No |
ExAC gnomAD |
|
|
rs770072024 COSM175453 COSM5074994 |
139 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs551009816 | 139 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 140 | P>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 142 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746261846 | 145 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs770297450 | 145 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs776751373 | 146 | T>N | No |
ExAC gnomAD |
|
| rs542068127 | 147 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs542068127 | 147 | T>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 148 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1805377471 | 148 | V>G | No | Ensembl | |
| rs1181973754 | 148 | V>M | No | TOPMed | |
| TCGA novel | 149 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1365141935 | 149 | A>T | No | gnomAD | |
| rs1316251675 | 152 | Q>* | No | gnomAD | |
| rs1362684299 | 152 | Q>H | No |
TOPMed gnomAD |
|
| rs1384001533 | 153 | S>N | No | gnomAD | |
| rs372053514 | 154 | F>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs150426007 | 155 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1805376601 | 156 | I>V | No | TOPMed | |
|
COSM5079469 rs141528182 |
157 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs779885233 | 157 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs779885233 | 157 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1805376267 | 158 | D>E | No | Ensembl | |
| rs1174065751 | 158 | D>Y | No | gnomAD | |
| rs966013135 | 159 | L>F | No |
TOPMed gnomAD |
|
|
rs4725617 VAR_028265 |
160 | V>A | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs4725617 | 160 | V>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs4725617 | 160 | V>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1008484548 | 161 | S>C | No | TOPMed | |
| rs1805375532 | 162 | G>S | No | gnomAD | |
| rs1194610095 | 162 | G>V | No | gnomAD | |
| TCGA novel | 163 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751226403 | 164 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs143405612 | 164 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs143405612 | 164 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs982934449 | 166 | L>P | No |
TOPMed gnomAD |
|
| rs763757809 | 167 | N>H | No |
ExAC gnomAD |
|
| rs762406186 | 167 | N>K | No |
ExAC gnomAD |
|
| rs1805374686 | 169 | E>D | No | TOPMed | |
| rs373364271 | 169 | E>K | No |
ESP ExAC gnomAD |
|
|
COSM3258329 rs199931138 |
170 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs199931138 | 170 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777199962 | 170 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs199931138 | 170 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs141275729 | 171 | C>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2116628550 | 172 | S>C | No | Ensembl | |
| rs1586584699 | 173 | L>R | No | Ensembl | |
| rs1377529661 | 175 | R>C | No |
TOPMed gnomAD |
|
| rs760180128 | 175 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs760180128 | 175 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs779038165 | 176 | L>V | No |
ExAC gnomAD |
|
| rs769688423 | 177 | T>P | No |
ExAC gnomAD |
|
|
COSM3878973 rs745810214 |
178 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs141392438 | 178 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs141392438 | 178 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs540355150 | 179 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs572755645 | 179 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 179 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs540355150 | 179 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1475956163 | 181 | L>F | No | gnomAD | |
| rs752203488 | 182 | Y>C | No |
ExAC gnomAD |
|
| rs1437597746 | 183 | L>P | No | gnomAD | |
|
COSM3411729 COSM3411730 rs1805372916 |
184 | A>T | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic TOPMed gnomAD |
| rs1805372845 | 184 | A>V | No | gnomAD | |
| rs1805372781 | 185 | F>L | No | Ensembl | |
| rs1198043030 | 187 | N>S | No | gnomAD | |
| rs766943140 | 188 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs766943140 | 188 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs375516618 | 189 | G>A | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs375516618 | 189 | G>D | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs1294128583 | 190 | A>D | No | gnomAD | |
| rs1423610614 | 191 | C>Y | No | gnomAD | |
| rs1441823997 | 192 | V>A | No | gnomAD | |
| rs1441823997 | 192 | V>G | No | gnomAD | |
| COSM5123474 | 193 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1805371888 | 194 | L>P | No | gnomAD | |
| rs774371662 | 195 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1162100202 | 197 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs781143121 | 198 | R>Q | No |
ExAC gnomAD |
|
| rs749318217 | 198 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2116628305 | 199 | V>G | No | Ensembl | |
| rs1805371266 | 199 | V>I | No | TOPMed | |
| rs1805371207 | 200 | F>S | No | gnomAD | |
| rs1231495693 | 201 | Y>* | No |
TOPMed gnomAD |
|
| rs557273137 | 201 | Y>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs557273137 | 201 | Y>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1212476853 | 202 | Q>L | No | TOPMed | |
| rs746832004 | 203 | R>C | No |
ExAC gnomAD |
|
| rs373761584 | 203 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs746832004 | 203 | R>S | No |
ExAC gnomAD |
|
| rs1805370683 | 206 | E>* | No |
TOPMed gnomAD |
|
| rs1805370683 | 206 | E>K | No |
TOPMed gnomAD |
|
| rs1337019566 | 209 | N>Y | No | gnomAD | |
| rs1805370142 | 210 | G>A | No | Ensembl | |
| rs201944315 | 210 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs778367248 | 212 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1281021546 | 212 | A>V | No |
TOPMed gnomAD |
|
| rs931862837 | 213 | Q>P | No | Ensembl | |
| rs754557302 | 216 | D>H | No |
ExAC gnomAD |
|
| rs754557302 | 216 | D>N | No |
ExAC gnomAD |
|
| rs1421420411 | 218 | L>P | No |
TOPMed gnomAD |
|
| rs1323793671 | 219 | P>S | No | gnomAD | |
| TCGA novel | 221 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753358941 | 221 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs756665403 | 222 | A>S | No |
ExAC TOPMed gnomAD |
|
|
rs756665403 COSM5176742 |
222 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1805369232 | 223 | G>E | No | TOPMed | |
| rs1563118730 | 226 | E>A | No | gnomAD | |
| rs921315527 | 226 | E>D | No | Ensembl | |
| rs1563118730 | 226 | E>V | No | gnomAD | |
| rs1398546577 | 227 | V>A | No | gnomAD | |
| rs1314564039 | 227 | V>M | No |
TOPMed gnomAD |
|
| rs1409728916 | 228 | A>S | No | gnomAD | |
| rs750821899 | 228 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs762026258 | 229 | G>E | No |
ExAC gnomAD |
|
| rs199988413 | 230 | T>A | No |
ExAC gnomAD |
|
| rs199988413 | 230 | T>P | No |
ExAC gnomAD |
|
| rs780968060 | 233 | P>T | No | Ensembl | |
| rs1215181856 | 234 | H>N | No | gnomAD | |
| rs1445436832 | 234 | H>P | No | gnomAD | |
| rs200473622 | 234 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1215181856 | 234 | H>Y | No | gnomAD | |
| rs770915770 | 235 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs3812408 | 235 | A>P | No |
TOPMed gnomAD |
|
| rs3812408 | 235 | A>T | No |
TOPMed gnomAD |
|
|
COSM3635275 rs770915770 |
235 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs747845357 | 236 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs747845357 | 236 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs201188496 | 236 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1586584398 | 237 | A>P | No | Ensembl | |
| rs754645542 | 239 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1805367092 | 239 | P>L | No |
TOPMed gnomAD |
|
| rs1805366945 | 241 | P>S | No | Ensembl | |
| rs779696210 | 242 | S>A | No |
ExAC gnomAD |
|
| rs912785967 | 243 | G>A | No |
TOPMed gnomAD |
|
| rs912785967 | 243 | G>D | No |
TOPMed gnomAD |
|
| rs912785967 | 243 | G>V | No |
TOPMed gnomAD |
|
| rs369211431 | 245 | P>H | No |
ESP TOPMed gnomAD |
|
| rs776794841 | 246 | R>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1455925427 | 246 | R>C | No |
TOPMed gnomAD |
|
|
COSM3878971 rs200235266 |
246 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs200235266 | 246 | R>L | No |
TOPMed gnomAD |
|
| rs1455925427 | 246 | R>S | No |
TOPMed gnomAD |
|
|
COSM1645247 COSM1645248 rs750956234 |
247 | M>I | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1412768892 | 249 | C>R | No |
TOPMed gnomAD |
|
| rs751843044 | 252 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs763151811 | 253 | G>A | No |
ExAC gnomAD |
|
| rs763151811 | 253 | G>V | No |
ExAC gnomAD |
|
| rs549085590 | 254 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs963454089 | 255 | W>C | No |
TOPMed gnomAD |
|
| rs1805364365 | 255 | W>R | No | TOPMed | |
| rs201380861 | 257 | V>G | No |
ExAC gnomAD |
|
| rs771860596 | 258 | P>A | No |
ExAC gnomAD |
|
| rs1805363595 | 258 | P>R | No | Ensembl | |
| rs771860596 | 258 | P>S | No |
ExAC gnomAD |
|
| rs748014434 | 259 | V>G | No | ExAC | |
| rs1805363372 | 259 | V>I | No | TOPMed | |
| rs1390169786 | 260 | G>E | No |
TOPMed gnomAD |
|
| COSM6176851 | 261 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201209720 | 261 | R>P | No |
1000Genomes TOPMed gnomAD |
|
| rs201209720 | 261 | R>Q | No |
1000Genomes TOPMed gnomAD |
|
|
rs140233341 COSM3878969 |
261 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1586584265 | 262 | C>G | No | gnomAD | |
| rs1586584265 | 262 | C>R | No | gnomAD | |
| rs1270306538 | 263 | H>Q | No | gnomAD | |
| rs1805362436 | 263 | H>R | No | Ensembl | |
| rs1266044572 | 265 | E>G | No |
TOPMed gnomAD |
|
| rs1353200268 | 265 | E>K | No | gnomAD | |
| rs748954306 | 266 | P>L | No |
ExAC TOPMed |
|
| rs768480626 | 266 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs779796093 | 267 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1805361597 | 267 | G>S | No | TOPMed | |
| rs755673712 | 268 | Y>D | No |
ExAC gnomAD |
|
| rs1450812457 | 269 | E>G | No |
TOPMed gnomAD |
|
| rs1189757406 | 269 | E>K | No | gnomAD | |
| rs1450812457 | 269 | E>V | No |
TOPMed gnomAD |
|
|
COSM3635273 rs745331095 |
270 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1264578895 | 271 | G>D | No | gnomAD | |
| rs781744273 | 272 | G>D | No |
ExAC gnomAD |
|
| rs757685325 | 274 | G>S | No |
ExAC TOPMed gnomAD |
|
|
COSM4681248 rs778110783 |
275 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs2116627508 | 276 | A>G | No | Ensembl | |
| rs1190425377 | 277 | C>S | No |
TOPMed gnomAD |
|
| rs998751117 | 278 | V>F | No | TOPMed | |
| rs754006853 | 279 | A>V | No |
ExAC gnomAD |
|
| rs1222426721 | 281 | P>S | No | gnomAD | |
| TCGA novel | 282 | S>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1047186045 | 283 | G>D | No |
TOPMed gnomAD |
|
| rs760751325 | 283 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1047186045 | 283 | G>V | No |
TOPMed gnomAD |
|
| rs751565118 | 284 | S>Y | No |
ExAC gnomAD |
|
| rs764084713 | 285 | Y>H | No |
ExAC gnomAD |
|
| rs149680322 | 286 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs759111320 | 286 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs149680322 | 286 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| COSM4941066 | 287 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2116626971 | 287 | M>T | No | Ensembl | |
| rs1586583888 | 288 | D>G | No | Ensembl | |
| rs139580332 | 289 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1420650893 | 289 | M>K | No | gnomAD | |
|
rs931346217 COSM373322 COSM373323 |
289 | M>V | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs376356192 | 292 | P>L | No |
ESP ExAC gnomAD |
|
| rs201804057 | 292 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201804057 | 292 | P>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1805352648 | 294 | C>F | No |
TOPMed gnomAD |
|
| rs372725643 | 294 | C>R | No |
ESP ExAC gnomAD |
|
| TCGA novel | 294 | C>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1805352648 | 294 | C>Y | No |
TOPMed gnomAD |
|
| rs1209926212 | 295 | L>F | No |
TOPMed gnomAD |
|
| rs749818737 | 296 | T>M | No |
ExAC gnomAD |
|
| rs2116626902 | 296 | T>P | No | Ensembl | |
| rs779130970 | 299 | Q>* | No |
ExAC gnomAD |
|
| rs1433233682 | 299 | Q>L | No | gnomAD | |
| rs1234156393 | 300 | Q>* | No | gnomAD | |
| rs1234156393 | 300 | Q>K | No | gnomAD | |
| rs1269104175 | 301 | S>I | No | gnomAD | |
| rs1179553928 | 301 | S>R | No | gnomAD | |
| rs1369463541 | 302 | T>A | No | gnomAD | |
| rs1305651240 | 303 | A>T | No | gnomAD | |
|
COSM599769 rs1805351654 COSM599770 |
306 | E>* | lung [Cosmic] | No |
cosmic curated TOPMed |
| rs755294761 | 307 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1287321074 | 308 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs150974578 | 308 | A>V | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs749476007 | 309 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1805351077 | 309 | T>I | No | Ensembl | |
| rs368306573 | 310 | I>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs368306573 | 310 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1199574359 | 313 | C>* | No |
TOPMed gnomAD |
|
| rs866186417 | 313 | C>F | No | Ensembl | |
| rs1805350772 | 313 | C>R | No | TOPMed | |
| rs55874355 | 314 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs538968659 COSM421566 COSM421565 |
314 | E>K | urinary_tract [Cosmic] | No |
cosmic curated 1000Genomes ExAC gnomAD |
| rs752606620 | 315 | S>N | No |
ExAC gnomAD |
|
| rs769788964 | 315 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs766012017 | 316 | G>D | No |
ExAC gnomAD |
|
| rs776167447 | 316 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1210636799 | 319 | R>G | No |
TOPMed gnomAD |
|
| rs1402059738 | 319 | R>S | No | Ensembl | |
| rs553135371 | 320 | A>G | No |
1000Genomes ExAC gnomAD |
|
| rs1346262028 | 320 | A>T | No | gnomAD | |
| rs553135371 | 320 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs1028814084 | 322 | G>R | No |
TOPMed gnomAD |
|
| rs1341375671 | 323 | E>D | No | gnomAD | |
| TCGA novel | 323 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1805348964 | 325 | P>S | No | TOPMed | |
| rs1805348666 | 326 | Q>H | No | TOPMed | |
| rs1563118204 | 326 | Q>P | No | Ensembl | |
| rs920137837 | 327 | V>A | No | TOPMed | |
| rs748648594 | 327 | V>M | No |
ExAC gnomAD |
|
| rs1377102797 | 329 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1787360676 | 330 | T>I | No | gnomAD | |
| rs1358763630 | 331 | G>D | No | gnomAD | |
| COSM3635271 | 331 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781511591 | 332 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1238073297 | 332 | P>L | No | gnomAD | |
| rs781511591 | 332 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs781511591 | 332 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs778751497 | 333 | P>L | No |
ExAC gnomAD |
|
| rs1008147879 | 333 | P>S | No |
TOPMed gnomAD |
|
| rs754919004 | 334 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs779818196 | 335 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs779818196 | 335 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs755838535 | 335 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1440878134 | 336 | P>H | No |
TOPMed gnomAD |
|
| rs1440878134 | 336 | P>L | No |
TOPMed gnomAD |
|
| rs1181013168 | 336 | P>S | No | gnomAD | |
| rs767116546 | 337 | R>* | No |
ExAC TOPMed gnomAD |
|
|
rs201581948 COSM48320 |
337 | R>Q | lung [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1805338938 | 338 | N>K | No | TOPMed | |
| rs1805338699 | 340 | S>R | No | Ensembl | |
| rs199837219 | 341 | F>C | No |
1000Genomes TOPMed |
|
| rs199837219 | 341 | F>S | No |
1000Genomes TOPMed |
|
| rs143872412 | 342 | S>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1805338501 | 342 | S>T | No | TOPMed | |
| rs1204837184 | 344 | S>P | No | gnomAD | |
| rs763514757 | 346 | T>A | No |
ExAC gnomAD |
|
| rs776076308 | 346 | T>I | No |
ExAC gnomAD |
|
| rs895369223 | 347 | Q>E | No |
TOPMed gnomAD |
|
| rs1240735868 | 347 | Q>R | No |
TOPMed gnomAD |
|
| rs1370189067 | 348 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1300542881 | 348 | L>P | No | gnomAD | |
| rs1056746694 | 349 | S>Y | No |
TOPMed gnomAD |
|
|
rs56006153 VAR_042115 |
351 | R>C | No |
UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM1205431 COSM1205432 rs111641464 |
351 | R>H | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs111641464 | 351 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1398632917 | 352 | W>C | No | gnomAD | |
| rs771244062 | 354 | P>T | No |
ExAC gnomAD |
|
| rs747104136 | 355 | P>T | No |
ExAC gnomAD |
|
| rs777893167 | 356 | A>E | No |
ExAC gnomAD |
|
| COSM4929245 | 357 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1192382687 | 357 | D>Y | No | gnomAD | |
| COSM4929218 | 358 | T>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs370810516 | 358 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1192707729 | 359 | G>A | No | gnomAD | |
| rs1192707729 | 359 | G>E | No | gnomAD | |
| rs750188342 | 360 | G>E | No |
ExAC gnomAD |
|
| rs139831060 | 360 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs200253721 | 361 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs144736263 COSM1086798 COSM1086799 |
361 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1367657243 | 362 | Q>H | No | gnomAD | |
| rs1220921084 | 362 | Q>P | No | gnomAD | |
| rs1220921084 | 362 | Q>R | No | gnomAD | |
| rs1269108743 | 364 | V>I | No |
TOPMed gnomAD |
|
| rs1478960763 | 365 | R>G | No | TOPMed | |
| rs1424510804 | 365 | R>S | No | TOPMed | |
| rs751098607 | 366 | Y>F | No |
ExAC gnomAD |
|
| rs965820153 | 366 | Y>H | No | Ensembl | |
| rs763667866 | 367 | S>T | No |
ExAC gnomAD |
|
| rs1805335025 | 370 | C>Y | No | TOPMed | |
| rs1420692583 | 371 | S>A | No | gnomAD | |
| rs1161570547 | 372 | Q>P | No |
TOPMed gnomAD |
|
| rs1161570547 | 372 | Q>R | No |
TOPMed gnomAD |
|
| rs1805334671 | 373 | C>Y | No | TOPMed | |
| rs765803133 | 374 | Q>* | No | ExAC | |
| rs1805334538 | 374 | Q>L | No | TOPMed | |
| COSM1086797 | 375 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs149370167 | 375 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| COSM1086795 | 376 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1474516797 | 376 | T>I | No | gnomAD | |
| rs1805334092 | 378 | Q>* | No | TOPMed | |
| rs964228335 | 379 | D>N | No |
TOPMed gnomAD |
|
| rs773238741 | 380 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs760945286 | 380 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs773238741 | 380 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1274962847 | 381 | G>E | No |
TOPMed gnomAD |
|
| rs1805333227 | 381 | G>R | No | Ensembl | |
| rs57652656 | 382 | P>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs57652656 | 382 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1212296171 | 385 | P>S | No | gnomAD | |
| rs1805332302 | 386 | C>W | No | Ensembl | |
| rs1034352541 | 387 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs150254405 | 387 | G>R | No |
ESP TOPMed gnomAD |
|
| rs756937821 | 389 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs145891509 | 390 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs145891509 COSM745166 COSM745165 |
390 | V>M | lung [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs752219228 | 391 | H>R | No |
ExAC gnomAD |
|
| rs1416363336 | 391 | H>Y | No |
TOPMed gnomAD |
|
| rs765890431 | 393 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1448845 COSM1448846 rs140236236 |
394 | P>L | liver large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs760889211 | 396 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs151135649 | 396 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs143087087 | 397 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs375682369 | 397 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs769791439 | 399 | L>F | No |
ExAC gnomAD |
|
| rs1483496235 | 400 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM4431866 | 400 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1041401654 | 401 | T>I | No | TOPMed | |
| rs776377613 | 402 | P>R | No |
ExAC gnomAD |
|
| rs745812223 | 402 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs745812223 | 402 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1805329865 | 403 | A>V | No | TOPMed | |
| rs1258191923 | 404 | V>L | No | gnomAD | |
| rs1805329541 | 405 | H>P | No | Ensembl | |
| rs373557157 | 405 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1332253649 | 405 | H>Y | No |
TOPMed gnomAD |
|
| rs1473289970 | 406 | V>A | No |
TOPMed gnomAD |
|
| rs1389778296 | 407 | N>D | No | gnomAD | |
| rs1230393947 | 407 | N>S | No |
TOPMed gnomAD |
|
| COSM3635264 | 408 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1412334805 | 412 | Y>H | No | TOPMed | |
| rs867325911 | 413 | A>V | No | Ensembl | |
| rs2116624872 | 414 | N>D | No | Ensembl | |
| rs1805328809 | 416 | T>N | No | TOPMed | |
| rs199888663 | 418 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1805328525 | 419 | V>A | No | Ensembl | |
| rs1388321070 | 419 | V>M | No | gnomAD | |
| TCGA novel | 420 | E>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778558358 | 420 | E>K | No |
ExAC gnomAD |
|
| rs766865327 | 421 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs754469465 | 421 | A>T | No |
ExAC gnomAD |
|
| rs766865327 | 421 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs942524237 | 422 | Q>* | No |
TOPMed gnomAD |
|
| rs942524237 | 422 | Q>E | No |
TOPMed gnomAD |
|
| rs756532762 | 425 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1805328001 | 426 | S>* | No | gnomAD | |
| rs767837551 | 427 | G>V | No |
ExAC gnomAD |
|
| rs1212536971 | 428 | L>Q | No |
TOPMed gnomAD |
|
| rs762019218 | 429 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs762019218 | 429 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1266841388 | 431 | S>C | No | gnomAD | |
| COSM421568 | 431 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1203283583 | 431 | S>P | No | TOPMed | |
| rs1322662465 | 432 | G>V | No |
TOPMed gnomAD |
|
| rs149068255 | 433 | H>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs368513616 | 434 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1440252878 | 436 | T>A | No | gnomAD | |
| rs763362632 | 436 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs763362632 | 436 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1440252878 | 436 | T>P | No | gnomAD | |
| rs1563117532 | 437 | S>L | No | Ensembl | |
| rs760562408 | 438 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs772865226 | 442 | M>V | No |
ExAC gnomAD |
|
| rs1805326512 | 443 | G>E | No | gnomAD | |
| rs771914513 | 445 | A>E | No |
ExAC gnomAD |
|
| rs1156638928 | 445 | A>T | No |
TOPMed gnomAD |
|
| rs747805007 | 446 | E>K | No |
ExAC gnomAD |
|
| rs773138765 | 450 | G>D | No |
ExAC gnomAD |
|
| rs1805320693 | 450 | G>S | No | Ensembl | |
| rs1805320167 | 451 | L>P | No | Ensembl | |
| rs774180152 | 452 | S>C | No |
ExAC gnomAD |
|
| rs774180152 | 452 | S>F | No |
ExAC gnomAD |
|
| rs768392833 | 456 | V>L | No |
ExAC gnomAD |
|
| rs748879034 | 457 | K>M | No |
ExAC gnomAD |
|
| rs1805319547 | 457 | K>N | No | TOPMed | |
| rs779424882 | 458 | K>T | No |
ExAC gnomAD |
|
| rs202178565 | 460 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1805319219 | 460 | P>S | No |
TOPMed gnomAD |
|
| rs1295746198 | 461 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs757242801 | 462 | Q>* | No | TOPMed | |
| rs1805318678 | 462 | Q>L | No | TOPMed | |
| rs1805318625 | 464 | E>G | No | Ensembl | |
| rs539891278 | 466 | T>N | No |
1000Genomes ExAC gnomAD |
|
| COSM1086793 | 467 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751886971 | 467 | W>L | No |
ExAC TOPMed gnomAD |
|
| rs757647389 | 467 | W>R | No |
ExAC TOPMed gnomAD |
|
| rs577729690 | 468 | A>V | No |
TOPMed gnomAD |
|
| rs758540262 | 469 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs1003754616 | 469 | G>R | No | TOPMed | |
| rs143535859 | 471 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145268229 | 471 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM1743259 rs201944531 COSM1743258 |
473 | R>* | Variant assessed as Somatic; HIGH impact. biliary_tract [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs201944531 | 473 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs147852131 | 473 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs147852131 | 473 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1331468050 | 474 | S>R | No |
TOPMed gnomAD |
|
| COSM1086791 | 475 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs187179716 | 475 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1187305212 | 476 | G>W | No | gnomAD | |
| rs373049955 | 477 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs775148876 | 480 | T>I | No |
ExAC gnomAD |
|
| rs1805316774 | 480 | T>P | No |
TOPMed gnomAD |
|
| rs769207409 | 481 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs73154214 | 484 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1805316262 | 485 | V>E | No | TOPMed | |
| rs144470322 | 485 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM1448844 COSM1448843 rs144470322 |
485 | V>M | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| TCGA novel | 487 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749437637 | 489 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs749437637 | 489 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs749437637 | 489 | D>Y | No |
ExAC TOPMed gnomAD |
|
|
rs11768549 VAR_028266 |
492 | R>Q | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM167118 COSM3411726 rs756203512 |
492 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine central_nervous_system [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1805309396 | 494 | Q>* | No |
TOPMed gnomAD |
|
| rs375144213 | 494 | Q>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 494 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752512314 | 495 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1303820676 | 495 | M>V | No |
TOPMed gnomAD |
|
| rs764851795 | 496 | V>A | No |
ExAC gnomAD |
|
| rs2116623305 | 496 | V>I | No | Ensembl | |
| rs759180125 | 497 | L>I | No |
ExAC gnomAD |
|
| rs759180125 | 497 | L>V | No |
ExAC gnomAD |
|
| rs1706447180 | 499 | P>L | No | Ensembl | |
| rs765891077 | 500 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs765891077 | 500 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1331331625 | 501 | V>F | No | gnomAD | |
| rs1399765615 | 507 | Q>* | No |
TOPMed gnomAD |
|
| rs1228564780 | 509 | D>N | No | Ensembl | |
| rs773742864 | 513 | I>V | No |
ExAC TOPMed gnomAD |
|
|
COSM320146 rs567483385 COSM320145 |
514 | V>I | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs567483385 | 514 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774766257 | 515 | R>* | No |
ExAC gnomAD |
|
| rs964910085 | 515 | R>S | No |
TOPMed gnomAD |
|
| rs1805307392 | 516 | V>F | No | TOPMed | |
|
rs200836382 COSM1644198 COSM1644199 |
517 | R>Q | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1193288958 | 518 | M>T | No | gnomAD | |
| rs372088764 | 518 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2116623121 | 520 | T>S | No | Ensembl | |
| rs371246344 | 524 | P>H | No | gnomAD | |
| rs371246344 | 524 | P>L | No | gnomAD | |
| rs1805306785 | 524 | P>S | No | Ensembl | |
| rs150816053 | 526 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1805306617 | 526 | P>S | No | TOPMed | |
| rs867003393 | 527 | F>I | No | Ensembl | |
| COSM452535 | 527 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3635258 | 528 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777617799 | 528 | S>Y | No |
ExAC gnomAD |
|
| COSM3635256 | 529 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1275810756 | 530 | D>N | No | gnomAD | |
| rs1449976693 | 531 | H>N | No | Ensembl | |
| rs1220447296 | 531 | H>R | No | gnomAD | |
| rs1181962967 | 532 | E>G | No | gnomAD | |
| rs778778160 | 534 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs142265364 | 534 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs765978732 | 536 | S>I | No |
ExAC TOPMed gnomAD |
|
| rs765978732 | 536 | S>N | No |
ExAC TOPMed gnomAD |
|
| COSM3635254 | 537 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1332849160 | 538 | P>A | No | gnomAD | |
| rs760220358 | 538 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs760220358 | 538 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1332849160 | 538 | P>S | No | gnomAD | |
| TCGA novel | 539 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1805296522 | 539 | V>A | No |
TOPMed gnomAD |
|
| rs771069813 | 540 | S>P | No |
ExAC gnomAD |
|
| rs774390706 | 542 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs747022455 | 542 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs774390706 | 542 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1277534592 | 545 | G>A | No |
TOPMed gnomAD |
|
| rs1805295704 | 545 | G>R | No | TOPMed | |
| TCGA novel | 547 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs936860521 | 547 | E>D | No | TOPMed | |
| rs1805295461 | 547 | E>K | No |
TOPMed gnomAD |
|
| rs1298929475 | 548 | I>T | No | gnomAD | |
| rs1381290223 | 549 | V>A | No |
TOPMed gnomAD |
|
| rs1805295088 | 550 | A>P | No | Ensembl | |
| rs779820434 | 550 | A>V | No |
ExAC gnomAD |
|
| rs1357284899 | 551 | V>D | No |
TOPMed gnomAD |
|
| rs535509887 | 551 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1425152855 | 552 | I>V | No |
TOPMed gnomAD |
|
| rs780668174 | 553 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs780668174 | 553 | F>Y | No |
ExAC TOPMed gnomAD |
|
| rs1207373721 | 555 | L>R | No |
TOPMed gnomAD |
|
| rs111778416 | 558 | G>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1805294004 | 558 | G>C | No | TOPMed | |
| rs111778416 | 558 | G>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM1661674 COSM1661675 rs1805294004 |
558 | G>S | kidney [Cosmic] | No |
cosmic curated TOPMed |
| rs111778416 | 558 | G>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1250670366 | 560 | A>G | No |
TOPMed gnomAD |
|
| rs1437436171 | 560 | A>T | No | TOPMed | |
| rs1805293117 | 564 | G>R | No | Ensembl | |
| TCGA novel | 566 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM86807 COSM3366837 rs555414886 |
567 | V>I | kidney ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs555414886 | 567 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1360684162 | 568 | F>S | No | gnomAD | |
| rs776947009 | 569 | R>P | No |
ExAC TOPMed gnomAD |
|
|
rs776947009 COSM3878959 |
569 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs200212760 | 569 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs765726381 | 570 | S>F | No |
TOPMed gnomAD |
|
| rs1365759219 | 571 | R>G | No | gnomAD | |
| rs1002258055 | 573 | A>T | No |
TOPMed gnomAD |
|
| rs560427865 | 573 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1366219382 | 574 | Q>* | No |
TOPMed gnomAD |
|
| rs1366219382 | 574 | Q>E | No |
TOPMed gnomAD |
|
| rs1263031167 | 574 | Q>H | No |
TOPMed gnomAD |
|
|
VAR_042116 rs35719334 |
575 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
UniProt NCI-TCGA TOPMed dbSNP gnomAD |
| rs780288031 | 575 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1301595315 | 576 | Q>H | No |
TOPMed gnomAD |
|
| rs1454467110 | 577 | R>* | No |
TOPMed gnomAD |
|
| rs750667620 | 577 | R>G | No |
ExAC gnomAD |
|
| rs1805283440 | 578 | Q>* | No | Ensembl | |
| rs1013254568 | 578 | Q>R | No | TOPMed | |
| rs1317538155 | 579 | Q>* | No | gnomAD | |
| rs1317538155 | 579 | Q>E | No | gnomAD | |
| rs1052212564 | 579 | Q>H | No | TOPMed | |
| rs1563116532 | 581 | Q>* | No | Ensembl | |
| rs574748801 | 582 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs555587376 | 582 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 582 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1229337821 | 583 | D>H | No | gnomAD | |
| rs983868627 | 584 | R>C | No |
TOPMed gnomAD |
|
| rs765284054 | 584 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
VAR_042117 COSM5152790 rs34178823 |
585 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic UniProt 1000Genomes NCI-TCGA TOPMed dbSNP gnomAD |
| rs1336834470 | 585 | A>V | No | gnomAD | |
| TCGA novel | 586 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1172447950 | 587 | D>E | No | gnomAD | |
| rs866192433 | 587 | D>H | No |
TOPMed gnomAD |
|
| rs866192433 | 587 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs866192433 | 587 | D>Y | No |
TOPMed gnomAD |
|
| rs1463832415 | 588 | V>M | No | gnomAD | |
| rs1374033123 | 589 | D>G | No | gnomAD | |
| rs1033504857 | 589 | D>N | No | TOPMed | |
| rs776941825 | 590 | R>* | No |
TOPMed gnomAD |
|
| rs776941825 | 590 | R>G | No |
TOPMed gnomAD |
|
| rs1052285437 | 590 | R>Q | No |
TOPMed gnomAD |
|
| rs1352423458 | 591 | E>D | No | TOPMed | |
| TCGA novel | 592 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781449260 | 592 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs781449260 | 592 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs369040226 | 593 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1221511235 | 594 | L>M | No |
TOPMed gnomAD |
|
| rs764091010 | 596 | L>M | No |
ExAC gnomAD |
|
| rs1805255051 | 597 | K>N | No | TOPMed | |
| rs758452852 | 598 | P>A | No |
ExAC gnomAD |
|
| rs753747935 | 599 | Y>* | No |
ExAC gnomAD |
|
| rs1805254747 | 600 | V>M | No | gnomAD | |
| rs1805254640 | 601 | D>N | No | Ensembl | |
| rs1805254383 | 603 | Q>H | No | TOPMed | |
| rs1419645656 | 604 | A>E | No | gnomAD | |
| rs1378490342 | 605 | Y>H | No | gnomAD | |
| rs754397131 | 606 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs146399527 | 607 | D>A | No |
ESP TOPMed gnomAD |
|
| rs1197085174 | 611 | G>* | No | gnomAD | |
| rs376643332 | 612 | A>D | No |
ESP TOPMed gnomAD |
|
| rs376643332 | 612 | A>G | No |
ESP TOPMed gnomAD |
|
| rs767349665 | 612 | A>T | No |
ExAC gnomAD |
|
| rs10952549 | 613 | L>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1678992035 | 613 | L>R | No | TOPMed | |
| TCGA novel | 614 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768075049 | 616 | T>S | No |
ExAC gnomAD |
|
| rs138935836 | 617 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs200018558 | 617 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1392328745 | 618 | E>Q | No | TOPMed | |
| rs1805251041 | 619 | L>I | No |
TOPMed gnomAD |
|
| rs1805251041 | 619 | L>V | No |
TOPMed gnomAD |
|
| rs1805250826 | 620 | D>G | No | Ensembl | |
| rs1284336251 | 620 | D>H | No | gnomAD | |
| rs1399859924 | 621 | P>L | No |
TOPMed gnomAD |
|
| rs1399859924 | 621 | P>R | No |
TOPMed gnomAD |
|
| rs200499248 | 622 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200499248 | 622 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1316479823 | 624 | L>R | No | TOPMed | |
| rs1190218089 | 625 | M>T | No |
TOPMed gnomAD |
|
| rs1805249902 | 626 | V>A | No | gnomAD | |
| rs747214273 | 627 | D>G | No |
ExAC gnomAD |
|
| rs1429662347 | 628 | T>A | No | gnomAD | |
| rs1805249359 | 628 | T>I | No | TOPMed | |
| rs1805249359 | 628 | T>S | No | TOPMed | |
| rs1196145922 | 629 | V>I | No |
TOPMed gnomAD |
|
| rs1476759571 | 630 | I>T | No | gnomAD | |
| rs1805248910 | 630 | I>V | No | TOPMed | |
| rs1805219347 | 633 | G>E | No |
TOPMed gnomAD |
|
| rs369094884 | 633 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 634 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs899153539 | 634 | E>D | No | TOPMed | |
| rs1170878071 | 637 | E>* | No |
TOPMed gnomAD |
|
| rs1404616704 | 637 | E>G | No | gnomAD | |
| rs1383290063 | 638 | V>L | No |
TOPMed gnomAD |
|
| rs544066979 | 639 | Y>C | No | gnomAD | |
|
COSM205472 rs112723648 |
640 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs112723648 | 640 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs761006248 | 640 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs529885927 | 641 | G>R | No |
1000Genomes ExAC gnomAD |
|
| rs1586579610 | 642 | T>P | No | Ensembl | |
|
COSM1645551 COSM1645552 rs1805218153 |
644 | R>K | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs772261312 | 645 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs1586579598 | 645 | L>P | No | Ensembl | |
| rs748417082 | 646 | P>L | No |
ExAC gnomAD |
|
| rs1805217669 | 647 | S>I | No | Ensembl | |
| rs768890787 | 648 | Q>* | No |
ExAC gnomAD |
|
| rs1290195084 | 649 | D>N | No | gnomAD | |
| rs1248856138 | 650 | C>R | No | gnomAD | |
| rs373965465 | 650 | C>S | No |
ESP TOPMed gnomAD |
|
| rs373965465 | 650 | C>Y | No |
ESP TOPMed gnomAD |
|
| rs749349744 | 651 | K>T | No |
ExAC gnomAD |
|
| rs2116616798 | 652 | T>A | No | Ensembl | |
| rs374663592 | 655 | I>M | No | Ensembl | |
| rs879098618 | 655 | I>V | No | TOPMed | |
|
rs1805216810 COSM4898074 |
658 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| rs781335770 | 659 | K>E | No |
ExAC gnomAD |
|
| rs1375790178 | 662 | S>Y | No |
TOPMed gnomAD |
|
| rs757213006 | 663 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs751386524 | 664 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs751386524 | 664 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1375071460 | 665 | G>S | No | TOPMed | |
| rs777636741 | 666 | Q>H | No |
ExAC gnomAD |
|
| rs993369891 | 666 | Q>R | No | TOPMed | |
| rs1805216048 | 667 | W>* | No | Ensembl | |
| rs1805216103 | 667 | W>R | No | TOPMed | |
| rs1805215988 | 670 | F>L | No | Ensembl | |
| rs1805215813 | 671 | L>F | No |
TOPMed gnomAD |
|
| COSM1086785 | 671 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758075798 | 672 | R>* | No |
ExAC gnomAD |
|
| rs752373855 | 672 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1398707074 | 673 | E>G | No |
TOPMed gnomAD |
|
| rs759092209 | 674 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs759092209 | 674 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs753335365 | 677 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs766864547 | 678 | G>R | No |
ExAC gnomAD |
|
| TCGA novel | 679 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2116616616 | 681 | S>C | No | 1000Genomes | |
| COSM3878957 | 682 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs140700329 | 683 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs200904525 | 683 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs774757061 | 684 | H>R | No |
ExAC gnomAD |
|
| rs1213712645 | 685 | I>V | No | gnomAD | |
| rs371479433 | 687 | H>L | No |
ExAC gnomAD |
|
| rs371479433 | 687 | H>P | No |
ExAC gnomAD |
|
| rs371479433 | 687 | H>R | No |
ExAC gnomAD |
|
| rs1805214243 | 687 | H>Y | No | gnomAD | |
| rs1343356019 | 688 | L>V | No | gnomAD | |
|
RCV000895356 rs145160235 |
691 | V>I | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs758249861 | 692 | V>I | No |
ExAC TOPMed gnomAD |
|
|
COSM1086781 COSM1086780 rs747914291 |
695 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| COSM3635252 | 695 | R>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1805209495 | 697 | P>A | No | Ensembl | |
|
rs34372369 VAR_042118 |
697 | P>L | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs752053540 | 699 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1364115555 | 701 | I>F | No | gnomAD | |
| rs1336934305 | 703 | E>D | No | gnomAD | |
| rs764574963 | 703 | E>G | No |
ExAC gnomAD |
|
| VAR_042119 | 703 | E>K | a breast pleomorphic lobular carcinoma sample; somatic mutation [UniProt] | No | UniProt |
| rs763368841 | 705 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1445606633 | 705 | M>V | No | gnomAD | |
| rs375566010 | 706 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1805208756 | 706 | E>V | No | TOPMed | |
| TCGA novel | 707 | N>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4947049 | 710 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 711 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 713 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1294092057 | 715 | R>W | No |
TOPMed gnomAD |
|
| rs147817243 | 716 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1222747501 | 717 | R>G | No | Ensembl | |
| rs144535927 | 717 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs144535927 | 717 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1222747501 | 717 | R>W | No | Ensembl | |
| rs758879954 | 718 | E>D | No |
ExAC gnomAD |
|
| rs1362250559 | 718 | E>K | No | gnomAD | |
| rs2116615669 | 719 | D>N | No | Ensembl | |
| rs548151465 | 720 | Q>* | No |
1000Genomes ExAC |
|
| rs149121680 | 724 | G>E | No |
ESP ExAC gnomAD |
|
| rs760710752 | 725 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1204021687 | 725 | Q>R | No | gnomAD | |
| rs774312704 | 727 | V>E | No |
ExAC gnomAD |
|
| rs768560146 | 729 | M>I | No |
ExAC gnomAD |
|
| rs1177683199 | 729 | M>R | No | TOPMed | |
| rs1283061958 | 731 | Q>K | No |
TOPMed gnomAD |
|
| rs775145198 | 733 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1805203620 | 733 | I>T | No | TOPMed | |
| rs1805203677 | 733 | I>V | No | Ensembl | |
| rs1367983073 | 734 | A>T | No | gnomAD | |
| rs553708553 | 737 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs915206166 | 739 | Y>* | No |
TOPMed gnomAD |
|
| rs1805203133 | 739 | Y>F | No | TOPMed | |
| rs1563115168 | 739 | Y>H | No | Ensembl | |
| rs964365496 | 741 | S>N | No |
TOPMed gnomAD |
|
| COSM3878955 | 741 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1805202930 | 742 | N>S | No | TOPMed | |
| rs1160462380 | 743 | H>Y | No |
TOPMed gnomAD |
|
| rs1454514009 | 744 | N>D | No | gnomAD | |
| rs1428259311 | 744 | N>S | No |
TOPMed gnomAD |
|
| rs1805202609 | 747 | H>Y | No | Ensembl | |
| rs145070507 | 748 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1086779 rs376954641 COSM1086778 |
748 | R>Q | endometrium [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs145070507 | 748 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs778478420 | 749 | D>H | No |
ExAC gnomAD |
|
| rs758966764 | 749 | D>V | No |
ExAC gnomAD |
|
| rs1205296887 | 751 | A>T | No | gnomAD | |
| rs1307412502 | 757 | V>E | No |
TOPMed gnomAD |
|
| rs1347822108 | 757 | V>M | No | gnomAD | |
| rs2116615370 | 758 | N>T | No | Ensembl | |
| rs1805201406 | 759 | Q>* | No | Ensembl | |
| rs1805201342 | 760 | N>K | No | gnomAD | |
|
COSM381177 rs1805201208 COSM381176 |
761 | L>Q | lung [Cosmic] | No |
cosmic curated Ensembl |
| rs1805201071 | 763 | C>R | No | TOPMed | |
| rs1758168775 | 763 | C>W | No | gnomAD | |
| rs756235783 | 763 | C>Y | No |
ExAC gnomAD |
|
| rs750490381 | 764 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs73726637 | 764 | K>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs73726637 | 764 | K>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1470816324 | 765 | V>L | No | gnomAD | |
| rs1425649832 | 766 | S>T | No | gnomAD | |
| rs775447726 | 767 | D>G | No |
ExAC gnomAD |
|
| rs1167212623 | 767 | D>N | No | gnomAD | |
| rs764971738 | 768 | F>S | No |
ExAC gnomAD |
|
| rs146094673 | 771 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1197422509 | 772 | R>C | No | TOPMed | |
| rs1197422509 | 772 | R>G | No | TOPMed | |
| rs550789024 | 772 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1012896062 | 773 | L>H | No |
TOPMed gnomAD |
|
| rs144262121 | 775 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs771554580 | 777 | F>C | No |
ExAC gnomAD |
|
| rs771554580 | 777 | F>S | No |
ExAC gnomAD |
|
| rs757458618 | 778 | D>E | No | Ensembl | |
| rs748628634 | 778 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs749718342 | 779 | G>D | No |
TOPMed gnomAD |
|
| rs373447921 | 779 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs749718342 | 779 | G>V | No |
TOPMed gnomAD |
|
| rs755342811 | 781 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs1382179005 | 781 | Y>H | No |
TOPMed gnomAD |
|
| rs1382179005 | 781 | Y>N | No |
TOPMed gnomAD |
|
|
COSM3258237 rs1454804041 |
782 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs549847780 | 785 | G>* | No |
ExAC TOPMed gnomAD |
|
| rs1490332858 | 786 | G>A | No |
TOPMed gnomAD |
|
| rs754947166 | 787 | K>R | No |
ExAC gnomAD |
|
| rs1805180693 | 789 | P>L | No |
TOPMed gnomAD |
|
| rs753768975 | 790 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs766301333 | 791 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs139711610 | 791 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs139711610 | 791 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM3832166 | 791 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2116614034 | 792 | W>C | No | Ensembl | |
| rs1805179812 | 793 | T>P | No | gnomAD | |
| rs1805179502 | 794 | A>G | No | Ensembl | |
| rs1805179605 | 794 | A>S | No | Ensembl | |
| rs1805179290 | 795 | P>S | No | TOPMed | |
| COSM1488316 | 796 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1805179198 | 797 | A>V | No | gnomAD | |
| rs767987164 | 798 | I>V | No |
ExAC gnomAD |
|
| rs1805178890 | 800 | H>R | No | gnomAD | |
| rs775933608 | 801 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs770181636 | 801 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs775933608 | 801 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1156441347 | 803 | F>C | No | gnomAD | |
| rs991771396 | 804 | T>A | No | TOPMed | |
| rs1805177911 | 805 | T>I | No | TOPMed | |
| rs781323096 | 806 | A>V | No | ExAC | |
| rs771180177 | 807 | S>C | No | ExAC | |
| COSM3878951 | 807 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
VAR_042120 rs56244405 |
807 | S>R | No |
UniProt TOPMed dbSNP gnomAD |
|
|
COSM745169 COSM745170 rs747132712 |
808 | D>N | lung Variant assessed as Somatic; MODERATE impact. skin [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1805177081 | 809 | V>M | No | Ensembl | |
| rs1193453669 | 810 | W>* | No |
TOPMed gnomAD |
|
| rs372075941 | 810 | W>* | No |
ESP ExAC gnomAD |
|
| rs1488827817 | 813 | G>R | No | gnomAD | |
| rs1805176623 | 814 | I>F | No | Ensembl | |
| rs758500982 | 816 | M>I | No |
ExAC gnomAD |
|
| rs753859464 | 817 | W>* | No |
ExAC gnomAD |
|
| TCGA novel | 818 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 818 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1586578240 | 819 | V>G | No | Ensembl | |
| rs780109666 | 819 | V>M | No |
ExAC gnomAD |
|
| rs1209190903 | 820 | L>P | No |
TOPMed gnomAD |
|
| rs1805175758 | 821 | S>G | No | TOPMed | |
| rs750219974 | 823 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1186505049 | 824 | D>E | No |
TOPMed gnomAD |
|
| rs761535126 | 826 | P>L | No |
ExAC gnomAD |
|
| rs767146354 | 826 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs767146354 | 826 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs190709674 | 828 | G>V | No | 1000Genomes | |
| rs1426261434 | 828 | G>W | No |
TOPMed gnomAD |
|
| rs763688617 | 830 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1164168455 | 830 | M>R | No |
TOPMed gnomAD |
|
| rs1164168455 | 830 | M>T | No |
TOPMed gnomAD |
|
| rs1175311514 | 831 | S>N | No | gnomAD | |
| rs1388224522 | 832 | N>S | No |
TOPMed gnomAD |
|
| rs1563114662 | 833 | Q>* | No |
TOPMed gnomAD |
|
| rs1805173824 | 834 | E>K | No | TOPMed | |
| rs1805163746 | 836 | M>T | No | gnomAD | |
| rs1008251277 | 838 | S>G | No | gnomAD | |
| rs1008251277 | 838 | S>R | No | gnomAD | |
| rs1362271571 | 839 | I>T | No |
TOPMed gnomAD |
|
| COSM3832164 | 840 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1390900021 | 840 | E>K | No | gnomAD | |
| rs951430650 | 841 | D>N | No | Ensembl | |
| rs764660537 | 842 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs764660537 | 842 | G>E | No |
ExAC TOPMed gnomAD |
|
|
COSM1205430 rs766796352 COSM1205429 |
844 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1086777 rs753822753 COSM1086776 |
844 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1213536991 | 846 | P>L | No | gnomAD | |
| rs555934791 | 847 | P>R | No |
1000Genomes ExAC gnomAD |
|
| COSM4902767 | 847 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772410524 | 849 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1805162328 | 852 | P>A | No | TOPMed | |
| rs748306167 | 853 | A>T | No |
ExAC gnomAD |
|
| rs768802916 | 854 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs774581597 | 854 | P>S | No |
ExAC gnomAD |
|
| rs2116612768 | 856 | Y>H | No | Ensembl | |
| rs1433805900 | 859 | M>R | No | gnomAD | |
| rs1433805900 | 859 | M>T | No | gnomAD | |
| rs1270475638 | 860 | K>N | No | TOPMed | |
| rs780958221 | 861 | N>K | No | ExAC | |
| COSM5164397 | 864 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1805161267 | 865 | Y>* | No | Ensembl | |
| rs1586577733 | 866 | D>A | No | Ensembl | |
| rs199929557 | 867 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs766464624 | 867 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs766464624 | 867 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1371180068 | 868 | A>D | No | gnomAD | |
| rs1371180068 | 868 | A>G | No | gnomAD | |
| rs758050233 | 869 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs1055875730 | 869 | R>H | No | Ensembl | |
| rs146374704 | 870 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs146374704 | 870 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs146374704 | 870 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs374097841 | 870 | R>W | No |
ESP TOPMed gnomAD |
|
| rs56124846 | 872 | H>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1418235400 | 873 | F>L | No | gnomAD | |
| rs1248159907 | 874 | Q>R | No | gnomAD | |
| rs1207144543 | 875 | K>E | No | gnomAD | |
| rs1483335998 | 876 | L>F | No | gnomAD | |
| rs1483335998 | 876 | L>I | No | gnomAD | |
| rs754403787 | 878 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs1805159716 | 878 | A>T | No |
TOPMed gnomAD |
|
| rs754411170 | 882 | Q>* | No |
ExAC gnomAD |
|
| rs891269774 | 884 | L>F | No |
TOPMed gnomAD |
|
| rs891269774 | 884 | L>I | No |
TOPMed gnomAD |
|
| rs537988053 | 888 | H>D | No |
ExAC gnomAD |
|
| rs537988053 | 888 | H>Y | No |
ExAC gnomAD |
|
| rs761160914 | 889 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs372390038 | 889 | S>P | No | gnomAD | |
| rs370603321 | 890 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs762143229 | 891 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs767930814 | 891 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs148388192 | 892 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs138978752 | 893 | I>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1164006815 | 893 | I>V | No | gnomAD | |
| rs1362341860 | 894 | A>S | No | gnomAD | |
| rs1362341860 | 894 | A>T | No | gnomAD | |
| rs776685006 | 895 | N>H | No |
ExAC gnomAD |
|
| rs770766668 | 896 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1472820846 | 896 | F>V | No | gnomAD | |
| rs1586577581 | 896 | F>Y | No | Ensembl | |
| rs746892705 | 897 | D>Y | No |
ExAC gnomAD |
|
| rs777558068 | 898 | P>H | No |
ExAC gnomAD |
|
| rs1248591499 | 898 | P>S | No |
TOPMed gnomAD |
|
| rs1764865478 | 899 | R>G | No | TOPMed | |
| rs778597320 | 899 | R>S | No |
ExAC gnomAD |
|
| rs1405011301 | 900 | M>I | No | gnomAD | |
| rs6967117 | 900 | M>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
VAR_028267 rs6967117 |
900 | M>V | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1586575815 | 901 | T>P | No | Ensembl | |
| TCGA novel | 902 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748905712 | 903 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs200265062 | 903 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748905712 | 903 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1167311375 | 906 | S>R | No | gnomAD | |
| rs1425745397 | 906 | S>T | No | gnomAD | |
| rs1805094085 | 907 | L>P | No | Ensembl | |
| rs1474111055 | 908 | S>G | No | gnomAD | |
| rs1391513028 | 908 | S>N | No | gnomAD | |
| TCGA novel | 909 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1188722292 | 912 | G>R | No | gnomAD | |
| rs755605986 | 913 | I>V | No |
ExAC TOPMed gnomAD |
|
|
COSM1086774 COSM1086775 rs751001344 |
914 | P>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs751001344 | 914 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1805093585 | 914 | P>S | No | Ensembl | |
| rs541812228 | 916 | R>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs541812228 | 916 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs919532275 COSM205471 |
916 | R>Q | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| TCGA novel | 918 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs530042951 | 918 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs759578750 | 920 | E>D | No |
ExAC gnomAD |
|
| rs146963652 | 920 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA |
| rs773180572 | 921 | W>C | No |
ExAC gnomAD |
|
| rs1554446949 | 923 | E>A | No |
TOPMed gnomAD |
|
| rs1554446949 | 923 | E>G | No |
TOPMed gnomAD |
|
| rs141196081 | 923 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs141196081 | 923 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748919069 | 924 | S>A | No | ExAC | |
| rs2116607287 | 925 | I>M | No | Ensembl | |
| rs1371801556 | 925 | I>T | No | gnomAD | |
| rs1427742183 | 925 | I>V | No | gnomAD | |
| rs138715519 | 926 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM74525 COSM4946952 rs138715519 |
926 | R>G | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC TOPMed gnomAD |
| rs201517649 | 926 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201517649 | 926 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs138715519 | 926 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs745352332 | 927 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1805090487 | 928 | K>E | No | TOPMed | |
| rs1586575626 | 928 | K>N | No | Ensembl | |
|
COSM1205435 rs781780617 COSM1205436 |
929 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs201365734 | 929 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 930 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747457918 | 931 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs1805089338 | 934 | F>S | No |
TOPMed gnomAD |
|
| rs1277477714 | 934 | F>V | No | gnomAD | |
| COSM1086773 | 934 | F>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752900413 | 935 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs765474060 | 936 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs1287929809 | 936 | S>A | No | gnomAD | |
|
COSM5651737 rs765474060 |
936 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1287929809 | 936 | S>P | No | gnomAD | |
| rs1407825756 | 938 | G>E | No |
TOPMed gnomAD |
|
| rs1805088030 | 938 | G>R | No | TOPMed | |
| rs1472955372 | 942 | M>K | No |
TOPMed gnomAD |
|
| rs1472955372 | 942 | M>R | No |
TOPMed gnomAD |
|
| rs753912953 | 942 | M>V | No |
ExAC gnomAD |
|
| rs1805087262 | 943 | E>D | No | gnomAD | |
| rs1162277611 | 943 | E>Q | No |
TOPMed gnomAD |
|
|
rs1308613660 COSM1205433 COSM1205434 |
944 | C>R | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs1423404234 | 944 | C>Y | No | gnomAD | |
| rs146015036 | 945 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1166389916 | 947 | E>D | No | gnomAD | |
| rs1805086597 | 947 | E>Q | No | TOPMed | |
| rs774299123 | 948 | L>P | No |
ExAC gnomAD |
|
| rs775057203 | 950 | A>D | No |
ExAC gnomAD |
|
| rs762781363 | 950 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs762781363 | 950 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM3258215 COSM216046 rs762781363 |
950 | A>T | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1805085858 | 951 | E>G | No | gnomAD | |
| rs1805085919 | 951 | E>K | No | TOPMed | |
| rs1805085919 | 951 | E>Q | No | TOPMed | |
| rs760208570 | 952 | D>G | No |
ExAC gnomAD |
|
| rs760208570 | 952 | D>V | No |
ExAC gnomAD |
|
| rs773648089 | 953 | L>Q | No | ExAC | |
| rs748538824 | 954 | T>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 955 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs142650387 | 957 | G>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs768961042 | 957 | G>R | No |
ExAC gnomAD |
|
| rs61732998 | 959 | T>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs61732998 | 959 | T>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1805080374 | 961 | P>L | No | TOPMed | |
| rs368297501 | 962 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1805079999 | 963 | H>N | No | TOPMed | |
| rs1805079688 | 965 | K>R | No |
TOPMed gnomAD |
|
|
rs753594142 COSM74524 COSM4947010 |
966 | R>C | ovary Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs139482378 | 966 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1186849061 | 967 | I>T | No | gnomAD | |
| rs1805079387 | 967 | I>V | No | TOPMed | |
| TCGA novel | 968 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs139639220 | 968 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs61732993 | 969 | C>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs61732993 | 969 | C>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1199635094 | 971 | I>T | No | gnomAD | |
|
rs771427284 COSM3635248 |
973 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA |
| rs1247231278 | 975 | K>E | No | TOPMed | |
| rs1482180615 | 975 | K>T | No |
TOPMed gnomAD |
|
| rs1805078324 | 976 | D>N | No | TOPMed | |
| rs1257638818 | 977 | D>C | No | gnomAD | |
| rs1805078241 | 977 | D>R | No | Ensembl |
No associated diseases with P21709
No regional properties for P21709
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P21709 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| fibronectin binding | Binding to a fibronectin, a group of related adhesive glycoproteins of high molecular weight found on the surface of animal cells, connective tissue matrices, and in extracellular fluids. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction |
| transmembrane-ephrin receptor activity | Combining with a transmembrane ephrin to initiate a change in cell activity. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of GTPase activity | Any process that initiates the activity of an inactive GTPase through the replacement of GDP by GTP. |
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| cell surface receptor signaling pathway | The series of molecular signals initiated by activation of a receptor on the surface of a cell. The pathway begins with binding of an extracellular ligand to a cell surface receptor, or for receptors that signal in the absence of a ligand, by ligand-withdrawal or the activity of a constitutively active receptor. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| negative regulation of cell migration | Any process that stops, prevents, or reduces the frequency, rate or extent of cell migration. |
| negative regulation of protein kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase activity. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| positive regulation of angiogenesis | Any process that activates or increases angiogenesis. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of cell-matrix adhesion | Any process that activates or increases the rate or extent of cell adhesion to an extracellular matrix. |
| positive regulation of stress fiber assembly | Any process that activates or increases the frequency, rate or extent of the assembly of a stress fiber, a bundle of microfilaments and other proteins found in fibroblasts. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| regulation of GTPase activity | Any process that modulates the rate of GTP hydrolysis by a GTPase. |
| substrate adhesion-dependent cell spreading | The morphogenetic process that results in flattening of a cell as a consequence of its adhesion to a substrate. |
88 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28693 | EPHB2 | Ephrin type-B receptor 2 | Gallus gallus (Chicken) | PR |
| Q07496 | EPHA4 | Ephrin type-A receptor 4 | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| Q07497 | EPHB5 | Ephrin type-B receptor 5 | Gallus gallus (Chicken) | PR |
| P29318 | EPHA3 | Ephrin type-A receptor 3 | Gallus gallus (Chicken) | SS |
| O42422 | EPHA7 | Ephrin type-A receptor 7 | Gallus gallus (Chicken) | SS |
| P54755 | EPHA5 | Ephrin type-A receptor 5 | Gallus gallus (Chicken) | SS |
| Q07498 | EPHB3 | Ephrin type-B receptor 3 | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| P29322 | EPHA8 | Ephrin type-A receptor 8 | Homo sapiens (Human) | SS |
| P54764 | EPHA4 | Ephrin type-A receptor 4 | Homo sapiens (Human) | SS |
| P54753 | EPHB3 | Ephrin type-B receptor 3 | Homo sapiens (Human) | SS |
| P29320 | EPHA3 | Ephrin type-A receptor 3 | Homo sapiens (Human) | PR |
| Q15375 | EPHA7 | Ephrin type-A receptor 7 | Homo sapiens (Human) | SS |
| Q5JZY3 | EPHA10 | Ephrin type-A receptor 10 | Homo sapiens (Human) | SS |
| P29323 | EPHB2 | Ephrin type-B receptor 2 | Homo sapiens (Human) | EV |
| Q9UF33 | EPHA6 | Ephrin type-A receptor 6 | Homo sapiens (Human) | SS |
| P54762 | EPHB1 | Ephrin type-B receptor 1 | Homo sapiens (Human) | SS |
| P54756 | EPHA5 | Ephrin type-A receptor 5 | Homo sapiens (Human) | SS |
| O15197 | EPHB6 | Ephrin type-B receptor 6 | Homo sapiens (Human) | SS |
| P07949 | RET | Proto-oncogene tyrosine-protein kinase receptor Ret | Homo sapiens (Human) | EV |
| O15146 | MUSK | Muscle, skeletal receptor tyrosine-protein kinase | Homo sapiens (Human) | EV |
| Q01974 | ROR2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Homo sapiens (Human) | EV |
| Q01973 | ROR1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Homo sapiens (Human) | PR |
| Q16832 | DDR2 | Discoidin domain-containing receptor 2 | Homo sapiens (Human) | SS |
| Q08345 | DDR1 | Epithelial discoidin domain-containing receptor 1 | Homo sapiens (Human) | EV |
| Q16620 | NTRK2 | BDNF/NT-3 growth factors receptor | Homo sapiens (Human) | EV |
| P04629 | NTRK1 | High affinity nerve growth factor receptor | Homo sapiens (Human) | EV |
| Q16288 | NTRK3 | NT-3 growth factor receptor | Homo sapiens (Human) | SS |
| P35916 | FLT4 | Vascular endothelial growth factor receptor 3 | Homo sapiens (Human) | SS |
| P17948 | FLT1 | Vascular endothelial growth factor receptor 1 | Homo sapiens (Human) | SS |
| P35968 | KDR | Vascular endothelial growth factor receptor 2 | Homo sapiens (Human) | EV |
| P29317 | EPHA2 | Ephrin type-A receptor 2 | Homo sapiens (Human) | SS |
| P54760 | EPHB4 | Ephrin type-B receptor 4 | Homo sapiens (Human) | SS |
| P29376 | LTK | Leukocyte tyrosine kinase receptor | Homo sapiens (Human) | SS |
| Q9UM73 | ALK | ALK tyrosine kinase receptor | Homo sapiens (Human) | EV |
| P08922 | ROS1 | Proto-oncogene tyrosine-protein kinase ROS | Homo sapiens (Human) | SS |
| P14616 | INSRR | Insulin receptor-related protein | Homo sapiens (Human) | SS |
| P08069 | IGF1R | Insulin-like growth factor 1 receptor | Homo sapiens (Human) | EV |
| P06213 | INSR | Insulin receptor | Homo sapiens (Human) | EV |
| P11362 | FGFR1 | Fibroblast growth factor receptor 1 | Homo sapiens (Human) | EV |
| P21802 | FGFR2 | Fibroblast growth factor receptor 2 | Homo sapiens (Human) | EV |
| P22455 | FGFR4 | Fibroblast growth factor receptor 4 | Homo sapiens (Human) | SS |
| P22607 | FGFR3 | Fibroblast growth factor receptor 3 | Homo sapiens (Human) | EV |
| Q12866 | MERTK | Tyrosine-protein kinase Mer | Homo sapiens (Human) | EV |
| Q04912 | MST1R | Macrophage-stimulating protein receptor | Homo sapiens (Human) | EV |
| P30530 | AXL | Tyrosine-protein kinase receptor UFO | Homo sapiens (Human) | PR |
| Q06418 | TYRO3 | Tyrosine-protein kinase receptor TYRO3 | Homo sapiens (Human) | SS |
| P08581 | MET | Hepatocyte growth factor receptor | Homo sapiens (Human) | EV |
| P04626 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | Homo sapiens (Human) | SS |
| Q15303 | ERBB4 | Receptor tyrosine-protein kinase erbB-4 | Homo sapiens (Human) | SS |
| P21860 | ERBB3 | Receptor tyrosine-protein kinase erbB-3 | Homo sapiens (Human) | SS |
| P00533 | EGFR | Epidermal growth factor receptor | Homo sapiens (Human) | EV |
| P34925 | RYK | Tyrosine-protein kinase RYK | Homo sapiens (Human) | PR |
| Q02763 | TEK | Angiopoietin-1 receptor | Homo sapiens (Human) | SS |
| P35590 | TIE1 | Tyrosine-protein kinase receptor Tie-1 | Homo sapiens (Human) | PR |
| P10721 | KIT | Mast/stem cell growth factor receptor Kit | Homo sapiens (Human) | EV |
| P09619 | PDGFRB | Platelet-derived growth factor receptor beta | Homo sapiens (Human) | EV |
| P07333 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Homo sapiens (Human) | SS |
| P16234 | PDGFRA | Platelet-derived growth factor receptor alpha | Homo sapiens (Human) | EV |
| P36888 | FLT3 | Receptor-type tyrosine-protein kinase FLT3 | Homo sapiens (Human) | EV |
| P54754 | Ephb3 | Ephrin type-B receptor 3 | Mus musculus (Mouse) | SS |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q03137 | Epha4 | Ephrin type-A receptor 4 | Mus musculus (Mouse) | SS |
| Q61772 | Epha7 | Ephrin type-A receptor 7 | Mus musculus (Mouse) | SS |
| O09127 | Epha8 | Ephrin type-A receptor 8 | Mus musculus (Mouse) | SS |
| Q8CBF3 | Ephb1 | Ephrin type-B receptor 1 | Mus musculus (Mouse) | SS |
| Q8BYG9 | Epha10 | Ephrin type-A receptor 10 | Mus musculus (Mouse) | SS |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| P29319 | Epha3 | Ephrin type-A receptor 3 | Mus musculus (Mouse) | SS |
| Q62413 | Epha6 | Ephrin type-A receptor 6 | Mus musculus (Mouse) | SS |
| P54763 | Ephb2 | Ephrin type-B receptor 2 | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| O08644 | Ephb6 | Ephrin type-B receptor 6 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P54757 | Epha5 | Ephrin type-A receptor 5 | Rattus norvegicus (Rat) | SS |
| P54759 | Epha7 | Ephrin type-A receptor 7 | Rattus norvegicus (Rat) | SS |
| P09759 | Ephb1 | Ephrin type-B receptor 1 | Rattus norvegicus (Rat) | SS |
| P0C0K7 | Ephb6 | Ephrin type-B receptor 6 | Rattus norvegicus (Rat) | SS |
| O08680 | Epha3 | Ephrin type-A receptor 3 | Rattus norvegicus (Rat) | SS |
| O61460 | vab-1 | Ephrin receptor 1 | Caenorhabditis elegans | SS |
| Q8RWZ5 | SD25 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94AG2 | SERK1 | Somatic embryogenesis receptor kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E991 | PRK6 | Pollen receptor-like kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FXF2 | RKF1 | Probable LRR receptor-like serine/threonine-protein kinase RFK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O13147 | ephb3 | Ephrin type-B receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O73878 | ephb4b | Ephrin type-B receptor 4b | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O13146 | epha3 | Ephrin type-A receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MERRWPLGLG | LVLLLCAPLP | PGARAKEVTL | MDTSKAQGEL | GWLLDPPKDG | WSEQQQILNG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TPLYMYQDCP | MQGRRDTDHW | LRSNWIYRGE | EASRVHVELQ | FTVRDCKSFP | GGAGPLGCKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TFNLLYMESD | QDVGIQLRRP | LFQKVTTVAA | DQSFTIRDLV | SGSVKLNVER | CSLGRLTRRG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LYLAFHNPGA | CVALVSVRVF | YQRCPETLNG | LAQFPDTLPG | PAGLVEVAGT | CLPHARASPR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PSGAPRMHCS | PDGEWLVPVG | RCHCEPGYEE | GGSGEACVAC | PSGSYRMDMD | TPHCLTCPQQ |
| 310 | 320 | 330 | 340 | 350 | 360 |
| STAESEGATI | CTCESGHYRA | PGEGPQVACT | GPPSAPRNLS | FSASGTQLSL | RWEPPADTGG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RQDVRYSVRC | SQCQGTAQDG | GPCQPCGVGV | HFSPGARGLT | TPAVHVNGLE | PYANYTFNVE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AQNGVSGLGS | SGHASTSVSI | SMGHAESLSG | LSLRLVKKEP | RQLELTWAGS | RPRSPGANLT |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YELHVLNQDE | ERYQMVLEPR | VLLTELQPDT | TYIVRVRMLT | PLGPGPFSPD | HEFRTSPPVS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RGLTGGEIVA | VIFGLLLGAA | LLLGILVFRS | RRAQRQRQQR | QRDRATDVDR | EDKLWLKPYV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| DLQAYEDPAQ | GALDFTRELD | PAWLMVDTVI | GEGEFGEVYR | GTLRLPSQDC | KTVAIKTLKD |
| 670 | 680 | 690 | 700 | 710 | 720 |
| TSPGGQWWNF | LREATIMGQF | SHPHILHLEG | VVTKRKPIMI | ITEFMENGAL | DAFLREREDQ |
| 730 | 740 | 750 | 760 | 770 | 780 |
| LVPGQLVAML | QGIASGMNYL | SNHNYVHRDL | AARNILVNQN | LCCKVSDFGL | TRLLDDFDGT |
| 790 | 800 | 810 | 820 | 830 | 840 |
| YETQGGKIPI | RWTAPEAIAH | RIFTTASDVW | SFGIVMWEVL | SFGDKPYGEM | SNQEVMKSIE |
| 850 | 860 | 870 | 880 | 890 | 900 |
| DGYRLPPPVD | CPAPLYELMK | NCWAYDRARR | PHFQKLQAHL | EQLLANPHSL | RTIANFDPRM |
| 910 | 920 | 930 | 940 | 950 | 960 |
| TLRLPSLSGS | DGIPYRTVSE | WLESIRMKRY | ILHFHSAGLD | TMECVLELTA | EDLTQMGITL |
| 970 | |||||
| PGHQKRILCS | IQGFKD |