P20339
Gene name |
RAB5A (RAB5) |
Protein name |
Ras-related protein Rab-5A |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5868 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
|
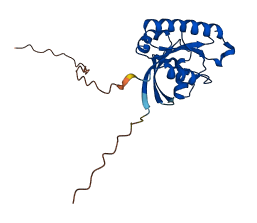
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

15 structures for P20339
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1N6H | X-ray | 151 A | A | 15-184 | PDB |
| 1N6I | X-ray | 160 A | A | 15-184 | PDB |
| 1N6K | X-ray | 155 A | A | 15-184 | PDB |
| 1N6L | X-ray | 160 A | A | 15-184 | PDB |
| 1N6N | X-ray | 160 A | A | 15-184 | PDB |
| 1N6O | X-ray | 180 A | A | 15-184 | PDB |
| 1N6P | X-ray | 154 A | A | 15-184 | PDB |
| 1N6R | X-ray | 155 A | A | 15-184 | PDB |
| 1R2Q | X-ray | 105 A | A | 15-184 | PDB |
| 1TU3 | X-ray | 231 A | A/B/C/D/E | 15-184 | PDB |
| 1TU4 | X-ray | 220 A | A/B/C/D | 15-184 | PDB |
| 3MJH | X-ray | 203 A | A/C | 16-183 | PDB |
| 4Q9U | X-ray | 462 A | B/F | 15-184 | PDB |
| 7BL1 | EM | 980 A | DDD | 16-183 | PDB |
| AF-P20339-F1 | Predicted | AlphaFoldDB |
143 variants for P20339
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA351870721 rs1575065947 |
2 | A>G | No |
ClinGen Ensembl |
|
|
CA2283947 rs774566349 |
3 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs200047397 CA351870756 |
4 | R>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2283948 rs200047397 |
4 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA351870768 rs1165832157 |
5 | G>D | No |
ClinGen gnomAD |
|
|
COSM2948363 rs144198452 CA2283950 |
6 | A>T | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA2283952 rs765282057 |
7 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765282057 CA351870796 |
7 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA351870814 rs1279277845 |
8 | R>G | No |
ClinGen TOPMed |
|
|
rs1450539066 CA351870854 |
10 | N>D | No |
ClinGen gnomAD |
|
|
rs1336388627 CA351870861 |
10 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA70736027 rs139006514 |
11 | G>E | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA351870871 rs1247949373 CA351870876 |
11 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA351870944 rs1160737451 |
14 | T>M | No |
ClinGen TOPMed |
|
|
CA2283955 rs149206559 |
14 | T>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs781668564 CA2283958 |
15 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1007656246 CA70736031 |
15 | G>R | No |
ClinGen TOPMed |
|
|
rs1202465417 CA351871025 |
17 | K>R | No |
ClinGen gnomAD |
|
|
CA2283960 rs147139196 |
19 | C>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA351871064 rs147139196 |
19 | C>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1175071032 CA351871072 |
19 | C>Y | No |
ClinGen gnomAD |
|
|
CA2283961 rs778287571 |
20 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA351871129 rs1418797273 |
22 | K>R | No |
ClinGen gnomAD |
|
| TCGA novel | 23 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 25 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2283967 rs772319638 |
37 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs772319638 CA2283966 |
37 | V>G | No |
ClinGen ExAC gnomAD |
|
|
CA351871486 rs1317673177 |
39 | R>C | No |
ClinGen TOPMed |
|
|
CA351871491 rs1559484871 |
39 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA351871533 rs1296605076 |
41 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs775197969 CA2283969 |
45 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs773130913 CA2283970 |
46 | H>N | No |
ClinGen ExAC gnomAD |
|
|
CA2283971 rs762827236 |
50 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1222784580 CA351871855 |
51 | S>N | No |
ClinGen gnomAD |
|
|
CA2283972 rs765895417 |
55 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs372972286 CA2284006 |
62 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs372972286 CA2284005 |
62 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs749099814 CA2284007 |
63 | C>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2284008 rs770773757 |
65 | D>A | No |
ClinGen ExAC gnomAD |
|
|
CA351857481 rs770773757 |
65 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs1468347379 CA351857514 |
67 | T>A | No |
ClinGen gnomAD |
|
|
rs745834147 CA351857576 |
70 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 74 | W>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 75 | D>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351857693 rs1458399542 |
76 | T>I | No |
ClinGen gnomAD |
|
|
rs1185033870 CA351857727 |
79 | Q>E | No |
ClinGen TOPMed |
|
|
CA351857725 rs1185033870 |
79 | Q>K | No |
ClinGen TOPMed |
|
|
CA351857732 rs1575076122 |
79 | Q>P | No |
ClinGen Ensembl |
|
|
rs1049089 CA70738526 |
81 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA70738530 rs969117779 COSM3365145 |
81 | R>Q | kidney [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1452938110 CA351857801 |
83 | H>R | No |
ClinGen gnomAD |
|
|
CA70738532 rs371989994 |
87 | P>L | No |
ClinGen ESP TOPMed |
|
|
rs760341655 CA2284013 |
88 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1440455566 CA351857855 |
91 | R>K | No |
ClinGen gnomAD |
|
|
rs1259412635 CA351857860 |
92 | G>R | No |
ClinGen TOPMed |
|
|
rs1198661606 CA351857870 |
93 | A>G | No |
ClinGen TOPMed |
|
|
CA70738540 rs896085692 |
94 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA351857879 rs549562506 |
95 | A>P | No |
ClinGen Ensembl |
|
|
rs549562506 CA70738542 |
95 | A>T | No |
ClinGen Ensembl |
|
|
CA351857881 rs1229696502 |
95 | A>V | No |
ClinGen TOPMed |
|
|
rs762022235 CA2284016 |
97 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1010513577 CA70738546 |
99 | V>L | No |
ClinGen TOPMed |
|
|
rs1034793274 CA70738548 |
101 | D>N | No |
ClinGen Ensembl |
|
| rs759467773 | 106 | E>= | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1324344650 CA351857966 |
106 | E>G | No |
ClinGen TOPMed |
|
|
CA2284039 rs752124120 |
106 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1559491993 CA351857972 |
107 | S>T | No |
ClinGen Ensembl |
|
|
CA2284041 rs373073333 |
109 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1184774975 CA351858009 |
111 | A>T | No |
ClinGen gnomAD |
|
|
rs1344123730 CA351858023 |
112 | K>E | No |
ClinGen gnomAD |
|
| TCGA novel | 113 | N>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351858125 rs1488785493 |
119 | Q>E | No |
ClinGen gnomAD |
|
|
rs756454883 CA2284043 |
119 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA2284046 rs181298476 |
123 | S>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs779797281 CA2284047 |
124 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs1451771428 CA351858199 |
124 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1451771428 CA351858201 |
124 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1383291378 CA351858204 |
125 | N>D | No |
ClinGen TOPMed |
|
|
rs140005569 CA2284048 |
125 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA70738655 rs375086132 |
125 | N>S | No |
ClinGen ESP gnomAD |
|
|
CA2284049 rs754401679 |
126 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1458883312 CA351858219 |
126 | I>V | No |
ClinGen gnomAD |
|
|
rs1575076324 CA351858253 |
128 | I>M | No |
ClinGen Ensembl |
|
|
rs1382973577 CA351858263 |
129 | A>G | No |
ClinGen gnomAD |
|
|
rs1321527286 CA351858289 |
131 | S>L | No |
ClinGen TOPMed gnomAD |
|
|
CA351858309 rs1450671486 |
133 | N>S | No |
ClinGen gnomAD |
|
|
CA70738663 rs550288787 |
135 | A>G | No |
ClinGen gnomAD |
|
|
CA2284054 rs749601586 |
136 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs760110691 CA2284057 |
138 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs760110691 CA2284058 |
138 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA2284059 rs775680725 |
142 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs1459723987 CA351858423 |
142 | A>T | No |
ClinGen gnomAD |
|
|
CA2284082 rs764274481 |
148 | A>P | No |
ClinGen ExAC gnomAD |
|
|
rs200242114 CA2284083 |
148 | A>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs201422065 CA2284084 |
149 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs766063435 CA2284085 |
151 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1341285683 CA351859243 |
151 | Y>H | No |
ClinGen TOPMed |
|
|
CA351859272 rs1274095230 |
152 | A>G | No |
ClinGen gnomAD |
|
|
CA351859268 rs1254575777 |
152 | A>S | No |
ClinGen gnomAD |
|
|
rs1190653315 CA351859282 |
153 | D>A | No |
ClinGen TOPMed gnomAD |
|
|
CA351859284 rs1190653315 |
153 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs751408634 CA2284086 |
153 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA351859334 rs1435148044 |
155 | N>S | No |
ClinGen gnomAD |
|
|
CA351859403 rs1369719292 |
160 | M>K | No |
ClinGen gnomAD |
|
|
CA70739377 rs147085379 CA2284087 |
160 | M>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA351859400 rs147085379 |
160 | M>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs772699440 CA2284088 |
161 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA2284091 rs777286944 |
164 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753467909 CA2284092 |
167 | S>* | No |
ClinGen ExAC gnomAD |
|
|
rs998497699 CA70739387 |
167 | S>T | No |
ClinGen Ensembl |
|
|
rs575707256 CA2284093 |
168 | M>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA351859532 rs1413976208 |
169 | N>K | No |
ClinGen TOPMed |
|
|
CA351859556 rs1559492752 |
171 | N>I | No |
ClinGen Ensembl |
|
|
CA351859607 rs1212448971 |
175 | M>V | No |
ClinGen gnomAD |
|
| TCGA novel | 176 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2284094 rs779142398 |
177 | I>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 180 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA351860488 rs1190693269 |
180 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1472341323 CA351860502 |
182 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
CA2284114 rs758749672 |
184 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs780583803 CA2284115 |
185 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA2284116 rs747492893 |
186 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1157810705 CA351860552 |
189 | P>L | No |
ClinGen gnomAD |
|
|
rs781415431 CA2284118 |
191 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748204519 CA2284119 |
193 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA2284120 rs769770592 |
194 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs55823411 CA70741468 |
195 | R>G | No |
ClinGen Ensembl |
|
|
rs1285345308 CA351860584 |
195 | R>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1382009772 CA351860604 |
198 | G>E | No |
ClinGen gnomAD |
|
|
CA351860607 rs1227048864 |
199 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA351860608 rs1227048864 |
199 | V>L | No |
ClinGen gnomAD |
|
|
rs150383454 CA70741471 |
203 | E>D | No |
ClinGen ESP |
|
|
CA2284123 rs771833104 |
203 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775305902 CA2284124 |
205 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA2284126 rs764079310 |
207 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA351860680 rs761318666 |
210 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2284128 rs761318666 |
210 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 211 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA70741493 rs764684048 |
211 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA351860686 rs1249748048 |
211 | Q>P | No |
ClinGen TOPMed |
|
|
CA351860707 rs1265203602 |
214 | S>G | No |
ClinGen TOPMed |
|
| TCGA novel | 215 | N>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs758032572 CA2284131 |
216 | N>Q | No |
ClinGen ExAC gnomAD |
No associated diseases with P20339
1 regional properties for P20339
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 21 - 176 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
29 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| anchored component of synaptic vesicle membrane | The component of the synaptic vesicle membrane consisting of the gene products that are tethered to the membrane only by a covalently attached anchor, such as a lipid group that is embedded in the membrane. Gene products with peptide sequences that are embedded in the membrane are excluded from this grouping. |
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| axon terminus | Terminal inflated portion of the axon, containing the specialized apparatus necessary to release neurotransmitters. The axon terminus is considered to be the whole region of thickening and the terminal button is a specialized region of it. |
| clathrin-coated endocytic vesicle membrane | The lipid bilayer surrounding a clathrin-coated endocytic vesicle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic side of early endosome membrane | The side (leaflet) of the early endosome membrane that faces the cytoplasm. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| early endosome | A membrane-bounded organelle that receives incoming material from primary endocytic vesicles that have been generated by clathrin-dependent and clathrin-independent endocytosis; vesicles fuse with the early endosome to deliver cargo for sorting into recycling or degradation pathways. |
| early endosome membrane | The lipid bilayer surrounding an early endosome. |
| early phagosome | A membrane-bounded intracellular vesicle as initially formed upon the ingestion of particulate material by phagocytosis. |
| endocytic vesicle | A membrane-bounded intracellular vesicle formed by invagination of the plasma membrane around an extracellular substance. Endocytic vesicles fuse with early endosomes to deliver the cargo for further sorting. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| melanosome | A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| phagocytic vesicle | A membrane-bounded intracellular vesicle that arises from the ingestion of particulate material by phagocytosis. |
| phagocytic vesicle membrane | The lipid bilayer surrounding a phagocytic vesicle. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic early endosome | An early endosome of the postsynapse. It acts as the major sorting station on the endocytic pathway, targeting neurotransmitter receptors for degregation or recycling. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
| somatodendritic compartment | The region of a neuron that includes the cell body (cell soma) and dendrite(s), but excludes the axon. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
| terminal bouton | Terminal inflated portion of the axon, containing the specialized apparatus necessary to release neurotransmitters. The axon terminus is considered to be the whole region of thickening and the terminal bouton is a specialized region of it. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein activity | A molecular function regulator that cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular processes. Intrinsic GTPase activity returns the G protein to its GDP-bound state. The return to the GDP-bound state can be accelerated by the action of a GTPase-activating protein (GAP). |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
15 GO annotations of biological process
| Name | Definition |
|---|---|
| amyloid-beta clearance by transcytosis | The process in which amyloid-beta is removed from extracellular brain regions by cell surface receptor-mediated endocytosis, followed by transcytosis across the blood-brain barrier. |
| early endosome to late endosome transport | The directed movement of substances, in membrane-bounded vesicles, from the early sorting endosomes to the late sorting endosomes; transport occurs along microtubules and can be experimentally blocked with microtubule-depolymerizing drugs. |
| endocytosis | A vesicle-mediated transport process in which cells take up external materials or membrane constituents by the invagination of a small region of the plasma membrane to form a new membrane-bounded vesicle. |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| modulation by host of viral process | A process in which a host organism modulates the frequency, rate or extent of any of a process being mediated by a virus with which it is infected. |
| phagocytosis | A vesicle-mediated transport process that results in the engulfment of external particulate material by phagocytes and their delivery to the lysosome. The particles are initially contained within phagocytic vacuoles (phagosomes), which then fuse with primary lysosomes to effect digestion of the particles. |
| positive regulation of exocytosis | Any process that activates or increases the frequency, rate or extent of exocytosis. |
| receptor internalization involved in canonical Wnt signaling pathway | A receptor internalization process that contributes to canonical Wnt signaling pathway. |
| regulation of autophagosome assembly | Any process that modulates the frequency, rate or extent of autophagosome assembly. |
| regulation of endocytosis | Any process that modulates the frequency, rate or extent of endocytosis. |
| regulation of endosome size | Any process that modulates the volume of an endosome, a membrane-bounded organelle that carries materials newly ingested by endocytosis. |
| regulation of filopodium assembly | Any process that modulates the frequency, rate or extent of the assembly of a filopodium, a thin, stiff protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal growth cone. |
| regulation of long-term neuronal synaptic plasticity | A process that modulates long-term neuronal synaptic plasticity, the ability of neuronal synapses to change long-term as circumstances require. Long-term neuronal synaptic plasticity generally involves increase or decrease in actual synapse numbers. |
| regulation of synaptic vesicle exocytosis | Any process that modulates the frequency, rate or extent of synaptic vesicle exocytosis. |
| synaptic vesicle recycling | The trafficking of synaptic vesicles from the pre-synaptic membrane so the vesicle can dock and prime for another round of exocytosis and neurotransmitter release. Recycling occurs after synaptic vesicle exocytosis, and is necessary to replenish presynaptic vesicle pools, sustain transmitter release and preserve the structural integrity of the presynaptic membrane. Recycling can occur following transient fusion with the presynaptic membrane (kiss and run), or via endocytosis of presynaptic membrane. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P38146 | YPT10 | GTP-binding protein YPT10 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q9NRW1 | RAB6B | Ras-related protein Rab-6B | Homo sapiens (Human) | PR |
| Q5JT25 | RAB41 | Ras-related protein Rab-41 | Homo sapiens (Human) | PR |
| P20340 | RAB6A | Ras-related protein Rab-6A | Homo sapiens (Human) | PR |
| Q9NP72 | RAB18 | Ras-related protein Rab-18 | Homo sapiens (Human) | PR |
| A4D1S5 | RAB19 | Ras-related protein Rab-19 | Homo sapiens (Human) | PR |
| Q9ULC3 | RAB23 | Ras-related protein Rab-23 | Homo sapiens (Human) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASRGATRPN | GPNTGNKICQ | FKLVLLGESA | VGKSSLVLRF | VKGQFHEFQE | STIGAAFLTQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TVCLDDTTVK | FEIWDTAGQE | RYHSLAPMYY | RGAQAAIVVY | DITNEESFAR | AKNWVKELQR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QASPNIVIAL | SGNKADLANK | RAVDFQEAQS | YADDNSLLFM | ETSAKTSMNV | NEIFMAIAKK |
| 190 | 200 | 210 | |||
| LPKNEPQNPG | ANSARGRGVD | LTEPTQPTRN | QCCSN |