P20338
Gene name |
RAB4A (RAB4) |
Protein name |
Ras-related protein Rab-4A |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5867 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
|
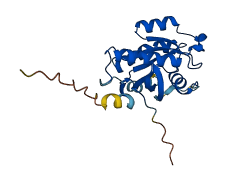
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for P20338
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1YU9 | X-ray | 207 A | A | 7-177 | PDB |
| 1Z0K | X-ray | 192 A | A/C | 7-177 | PDB |
| 2BMD | X-ray | 180 A | A | 6-189 | PDB |
| 2BME | X-ray | 157 A | A/B/C/D | 6-189 | PDB |
| AF-P20338-F1 | Predicted | AlphaFoldDB |
146 variants for P20338
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA38756531 rs369909116 |
3 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA345119986 rs1471211335 |
4 | T>M | No |
ClinGen gnomAD |
|
|
CA345119994 rs1335151723 |
5 | A>T | No |
ClinGen gnomAD |
|
|
rs1169802979 CA345120048 |
7 | S>T | No |
ClinGen TOPMed |
|
|
rs902791555 CA38756544 |
8 | E>K | No |
ClinGen TOPMed |
|
|
CA1442283 rs201255535 |
12 | F>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA345124096 rs1287786891 |
20 | G>V | No |
ClinGen gnomAD |
|
|
rs1337124947 CA345124108 |
21 | N>D | No |
ClinGen TOPMed |
|
|
CA345124128 rs1406721069 |
22 | A>T | No |
ClinGen gnomAD |
|
|
rs1286533669 CA345124173 |
24 | T>I | No |
ClinGen TOPMed |
|
|
rs1344711456 CA345124236 |
28 | C>F | No |
ClinGen gnomAD |
|
|
rs776521010 CA1442287 |
29 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1286011944 CA345124386 |
35 | E>G | No |
ClinGen gnomAD |
|
| TCGA novel | 35 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781323910 | 37 | K>N | Variant assessed as Somatic; 4.846e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1469927600 CA345124737 |
38 | F>L | No |
ClinGen gnomAD |
|
|
rs1558237148 CA345124449 |
38 | F>L | No |
ClinGen Ensembl |
|
|
rs1176333732 CA345124749 |
39 | K>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA345124762 rs1385458366 |
40 | D>N | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 44 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1161134692 CA345124871 |
45 | T>A | No |
ClinGen gnomAD |
|
| TCGA novel | 45 | T>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1278944601 CA345124904 |
46 | I>T | No |
ClinGen TOPMed |
|
|
CA1442307 rs180963002 |
46 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA345124928 rs1352590783 |
48 | V>M | No |
ClinGen gnomAD |
|
|
rs1034320973 CA38766755 |
49 | E>* | No |
ClinGen Ensembl |
|
|
rs1441206040 CA345125019 |
51 | G>D | No |
ClinGen gnomAD |
|
|
rs1397032625 CA345125064 |
54 | I>V | No |
ClinGen TOPMed |
|
|
CA345125119 rs1314499748 |
56 | N>T | No |
ClinGen gnomAD |
|
|
CA1442311 rs773328557 |
57 | V>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs36017250 CA1442312 |
58 | G>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs766522960 CA1442313 |
58 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs542539411 CA1442315 |
60 | K>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1442317 rs752998244 |
62 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1461929785 CA345125221 |
63 | K>N | No |
ClinGen TOPMed |
|
| TCGA novel | 67 | W>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 71 | G>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA345125465 rs1475436090 |
74 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs754296333 CA1442323 |
75 | F>L | No |
ClinGen ExAC |
|
|
CA38766832 rs866558785 |
76 | R>G | No |
ClinGen Ensembl |
|
|
rs1252542011 CA345126929 |
77 | S>Y | No |
ClinGen gnomAD |
|
|
rs141754859 CA1442339 |
78 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1172272102 CA345126946 |
79 | T>M | No |
ClinGen gnomAD |
|
|
rs1402597957 CA345126950 |
80 | R>G | No |
ClinGen gnomAD |
|
|
rs779534597 CA1442342 |
84 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs532123253 CA1442343 |
84 | R>Q | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1442345 rs780586276 |
86 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA345127065 rs1471039600 |
86 | A>V | No |
ClinGen gnomAD |
|
|
rs1206488700 CA345127108 |
88 | G>A | No |
ClinGen gnomAD |
|
|
rs777649734 CA1442348 |
88 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1442349 rs749151689 |
90 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 91 | L>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1364117608 CA345127178 |
92 | V>A | No |
ClinGen gnomAD |
|
|
CA1442351 rs774386684 |
92 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs139167201 CA1442353 |
93 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs199897325 CA1442354 |
94 | D>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1442355 rs11544296 |
96 | T>P | No |
ClinGen ExAC gnomAD |
|
| rs1281220421 | 97 | S>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1442374 rs768864729 |
98 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs776882217 CA1442375 |
98 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs765592653 CA1442377 |
100 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1571835049 CA345127928 |
101 | Y>C | No |
ClinGen Ensembl |
|
|
CA1442378 rs773449580 |
102 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1390025426 CA345127994 |
103 | A>E | No |
ClinGen gnomAD |
|
|
rs1324026251 CA345127992 |
103 | A>T | No |
ClinGen TOPMed |
|
|
COSM905229 rs1390025426 CA345128003 |
103 | A>V | Variant assessed as Somatic; impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1558240682 CA345128030 |
105 | T>A | No |
ClinGen Ensembl |
|
|
CA1442380 rs767047430 |
112 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345128241 rs1246206281 |
112 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA1442381 rs752249758 |
114 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755723609 CA1442382 |
115 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1478020729 CA345128344 |
116 | S>N | No |
ClinGen TOPMed |
|
|
CA1442385 rs756892543 |
119 | I>T | No |
ClinGen ExAC |
|
|
rs753491951 CA1442384 |
119 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1193866538 CA345128478 |
121 | I>L | No |
ClinGen gnomAD |
|
|
rs1193866538 CA345128485 |
121 | I>V | No |
ClinGen gnomAD |
|
|
rs1237275056 CA345128541 |
123 | L>F | No |
ClinGen gnomAD |
|
|
rs1311292750 CA345128553 |
123 | L>R | No |
ClinGen gnomAD |
|
|
CA345128758 rs1423288791 |
131 | D>N | No |
ClinGen gnomAD |
|
|
CA345128806 rs1378551658 |
132 | A>T | No |
ClinGen TOPMed |
|
|
rs996238062 CA345128847 |
134 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs996238062 CA38770996 |
134 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs370982346 CA1442388 COSM1626882 |
134 | R>H | Variant assessed as Somatic; 0.0 impact. liver [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs370982346 CA1442389 |
134 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370982346 CA1442387 |
134 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1442390 rs747062264 |
141 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA38771025 rs763779125 |
141 | A>V | No |
ClinGen Ensembl |
|
|
CA345129034 rs1215181509 |
142 | S>A | No |
ClinGen TOPMed |
|
|
CA345129040 rs1452976516 |
142 | S>C | No |
ClinGen gnomAD |
|
|
rs561383857 CA38771026 |
143 | R>G | No |
ClinGen 1000Genomes |
|
|
CA1442391 rs768777080 |
144 | F>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA38771027 rs1031302738 |
144 | F>L | No |
ClinGen TOPMed |
|
|
rs900760752 CA38771030 |
145 | A>T | No |
ClinGen Ensembl |
|
|
rs1217389690 CA345129127 |
145 | A>V | No |
ClinGen TOPMed |
|
|
rs776718032 CA1442392 |
146 | Q>* | No |
ClinGen ExAC |
|
| TCGA novel | 147 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA38771040 rs748669860 |
148 | N>H | No |
ClinGen Ensembl |
|
|
CA1442414 rs770048438 |
149 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1442415 rs777896179 |
151 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs749639801 CA1442416 |
152 | F>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM679196 rs774886245 CA1442418 |
157 | A>V | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA345130804 rs1571835836 |
162 | N>S | No |
ClinGen Ensembl |
|
|
CA345130813 rs1328639709 |
163 | V>I | No |
ClinGen TOPMed |
|
|
CA345130830 rs1424856330 |
164 | E>Q | No |
ClinGen gnomAD |
|
|
CA1442422 rs761298547 |
165 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs764915249 CA1442423 |
166 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs142406743 CA1442424 |
167 | F>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345130952 rs1179069825 |
169 | Q>* | No |
ClinGen TOPMed |
|
|
CA1442425 rs762619546 |
169 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345131041 COSM210255 rs1416962296 |
171 | A>T | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
| TCGA novel | 172 | R>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 173 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs766126067 CA1442426 CA345131185 |
176 | N>K | No |
ClinGen ExAC gnomAD |
|
|
CA345131172 rs1185359632 |
176 | N>T | No |
ClinGen TOPMed |
|
| TCGA novel | 177 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs781054509 CA1442429 |
179 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1252670098 CA345132809 |
182 | E>G | No |
ClinGen TOPMed |
|
|
rs1308507919 CA345132828 |
183 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1289792557 CA345132821 |
183 | L>V | No |
ClinGen gnomAD |
|
|
rs888851409 CA38773426 |
184 | D>H | No |
ClinGen gnomAD |
|
|
CA1442446 rs184851755 |
185 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1177599955 CA345132889 |
186 | E>G | No |
ClinGen gnomAD |
|
|
CA1442448 rs759364022 |
187 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1439238668 CA345132956 |
190 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs887541200 CA38773452 |
195 | G>R | No |
ClinGen TOPMed |
|
| TCGA novel | 196 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs151316203 CA1442450 |
196 | D>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 197 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1172306376 CA345133141 |
203 | R>G | No |
ClinGen gnomAD |
|
|
CA345133178 rs1397784625 COSM905231 |
205 | P>L | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs374069365 CA1442453 |
206 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1442452 rs764013458 |
206 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1442455 rs779345299 |
207 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA1442456 COSM905232 rs368207169 |
207 | R>H | large_intestine endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
COSM76151 CA1442458 rs200242732 |
208 | A>T | ovary Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA345133234 rs1317644451 |
209 | Q>R | No |
ClinGen gnomAD |
|
|
rs1263360422 CA345133251 |
210 | A>T | No |
ClinGen gnomAD |
|
|
CA1442459 rs747437276 |
210 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA1442460 rs754919585 |
211 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA38773513 rs754919585 |
211 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1442464 COSM679194 rs202214923 |
213 | A>S | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs202214923 CA1442463 |
213 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1421264360 CA345133300 |
213 | A>V | No |
ClinGen gnomAD |
|
|
CA345133301 rs1486473499 |
214 | Q>* | No |
ClinGen TOPMed |
|
|
CA1442465 COSM905234 rs759206677 |
217 | G>V | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1412915834 CA345133396 |
218 | C>G | No |
ClinGen gnomAD |
No associated diseases with P20338
1 regional properties for P20338
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 15 - 167 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasmic vesicle membrane | The lipid bilayer surrounding a cytoplasmic vesicle. |
| early endosome membrane | The lipid bilayer surrounding an early endosome. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| insulin-responsive compartment | A small membrane-bounded vesicle that releases its contents by exocytosis in response to insulin stimulation; the contents are enriched in GLUT4, IRAP and VAMP2. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic recycling endosome | A recycling endosome of the postsynapse. In postsynaptic terminals with dendritic spines, it is typically located at the base of a dendritic spine. It is involved in recycling of neurotransmitter receptors to the postsynaptic membrane. In some cases at least, this recycling is activated by postsynaptic signalling and so can play a role in long term potentiation. |
| recycling endosome | An organelle consisting of a network of tubules that functions in targeting molecules, such as receptors transporters and lipids, to the plasma membrane. |
| recycling endosome membrane | The lipid bilayer surrounding a recycling endosome. |
| vesicle | Any small, fluid-filled, spherical organelle enclosed by membrane. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein activity | A molecular function regulator that cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular processes. Intrinsic GTPase activity returns the G protein to its GDP-bound state. The return to the GDP-bound state can be accelerated by the action of a GTPase-activating protein (GAP). |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| antigen processing and presentation | The process in which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| Rab protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Rab family of proteins switching to a GTP-bound active state. |
| regulation of endocytosis | Any process that modulates the frequency, rate or extent of endocytosis. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2TBH7 | RAB4A | Ras-related protein Rab-4A | Bos taurus (Bovine) | PR |
| Q8WUD1 | RAB2B | Ras-related protein Rab-2B | Homo sapiens (Human) | PR |
| P61019 | RAB2A | Ras-related protein Rab-2A | Homo sapiens (Human) | PR |
| P61018 | RAB4B | Ras-related protein Rab-4B | Homo sapiens (Human) | PR |
| P62491 | RAB11A | Ras-related protein Rab-11A | Homo sapiens (Human) | PR |
| Q15907 | RAB11B | Ras-related protein Rab-11B | Homo sapiens (Human) | PR |
| Q91ZR1 | Rab4b | Ras-related protein Rab-4B | Mus musculus (Mouse) | PR |
| P51146 | Rab4b | Ras-related protein Rab-4B | Rattus norvegicus (Rat) | PR |
| P05714 | Rab4a | Ras-related protein Rab-4A | Rattus norvegicus (Rat) | PR |
| Q68EK7 | rab4b | Ras-related protein Rab-4B | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q6PHI9 | rab4a | Ras-related protein Rab-4A | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSQTAMSETY | DFLFKFLVIG | NAGTGKSCLL | HQFIEKKFKD | DSNHTIGVEF | GSKIINVGGK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YVKLQIWDTA | GQERFRSVTR | SYYRGAAGAL | LVYDITSRET | YNALTNWLTD | ARMLASQNIV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IILCGNKKDL | DADREVTFLE | ASRFAQENEL | MFLETSALTG | ENVEEAFVQC | ARKILNKIES |
| 190 | 200 | 210 | |||
| GELDPERMGS | GIQYGDAALR | QLRSPRRAQA | PNAQECGC |