P20226
Gene name |
TBP (GTF2D1, TF2D, TFIID) |
Protein name |
TATA-box-binding protein |
Names |
TATA sequence-binding protein, TATA-binding factor, TATA-box factor, Transcription initiation factor TFIID TBP subunit |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6908 |
EC number |
|
Protein Class |
TATA-BOX BINDING PROTEIN (PTHR10126) |
Descriptions
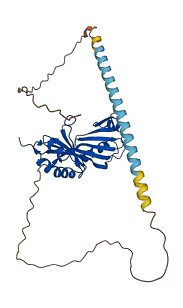
The TATA binding protein (TBP) is required for the expression of nearly all genes and is highly regulated both positively and negatively. Autoinhibition of TBP through dimerization contributes to transcriptional repression. In the absence of DNA, the conserved core of TBP crystallizes as a dimer, which occludes its DNA binding concave surface to prevent unregulated gene expression.
Autoinhibitory domains (AIDs)
Target domain |
159-338 (TATA binding domain) |
Relief mechanism |
Partner binding, Others |
Assay |
Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
References
- Chasman DI et al. (1993) "Crystal structure of yeast TATA-binding protein and model for interaction with DNA", Proceedings of the National Academy of Sciences of the United States of America, 90, 8174-8
- Jackson-Fisher AJ et al. (1999) "A role for TBP dimerization in preventing unregulated gene expression", Molecular cell, 3, 717-27
- Chitikila C et al. (2002) "Interplay of TBP inhibitors in global transcriptional control", Molecular cell, 10, 871-82
Autoinhibited structure

Activated structure

66 structures for P20226
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1C9B | X-ray | 265 A | B/F/J/N/R | 159-337 | PDB |
| 1CDW | X-ray | 190 A | A | 159-337 | PDB |
| 1JFI | X-ray | 262 A | C | 159-339 | PDB |
| 1NVP | X-ray | 210 A | A | 159-339 | PDB |
| 1TGH | X-ray | 290 A | A | 156-339 | PDB |
| 4ROC | X-ray | 190 A | B | 159-339 | PDB |
| 4ROD | X-ray | 270 A | B | 159-339 | PDB |
| 4ROE | X-ray | 220 A | B | 159-339 | PDB |
| 5FUR | EM | 850 A | A | 1-339 | PDB |
| 5IY6 | EM | 720 A | P | 1-339 | PDB |
| 5IY7 | EM | 860 A | P | 1-339 | PDB |
| 5IY8 | EM | 790 A | P | 1-339 | PDB |
| 5IY9 | EM | 630 A | P | 1-339 | PDB |
| 5IYA | EM | 540 A | P | 1-339 | PDB |
| 5IYB | EM | 390 A | P | 1-339 | PDB |
| 5IYC | EM | 390 A | P | 1-339 | PDB |
| 5IYD | EM | 390 A | P | 1-339 | PDB |
| 5N9G | X-ray | 270 A | B/G | 159-339 | PDB |
| 6MZD | EM | 980 A | T | 1-339 | PDB |
| 6MZL | EM | 2300 A | T | 1-339 | PDB |
| 6MZM | EM | 750 A | T | 1-339 | PDB |
| 6O9L | EM | 720 A | P | 1-339 | PDB |
| 7EDX | EM | 450 A | P | 1-339 | PDB |
| 7EG7 | EM | 620 A | P | 1-339 | PDB |
| 7EG8 | EM | 740 A | P | 1-339 | PDB |
| 7EG9 | EM | 370 A | P | 1-339 | PDB |
| 7EGA | EM | 410 A | P | 1-339 | PDB |
| 7EGB | EM | 330 A | P | 1-339 | PDB |
| 7EGC | EM | 390 A | P | 1-339 | PDB |
| 7EGD | EM | 675 A | P | 1-339 | PDB |
| 7EGE | EM | 900 A | P | 1-339 | PDB |
| 7EGF | EM | 316 A | P | 1-339 | PDB |
| 7EGI | EM | 982 A | P | 1-339 | PDB |
| 7EGJ | EM | 864 A | P | 1-339 | PDB |
| 7ENA | EM | 407 A | DP | 1-339 | PDB |
| 7ENC | EM | 413 A | DP | 1-339 | PDB |
| 7LBM | EM | 480 A | P | 1-339 | PDB |
| 7NVR | EM | 450 A | O | 1-339 | PDB |
| 7NVS | EM | 280 A | O | 1-339 | PDB |
| 7NVT | EM | 290 A | O | 1-339 | PDB |
| 7NVU | EM | 250 A | O | 1-339 | PDB |
| 7NVY | EM | 730 A | O | 1-339 | PDB |
| 7NVZ | EM | 720 A | O | 1-339 | PDB |
| 7NW0 | EM | 660 A | O | 1-339 | PDB |
| 7ZWC | EM | 320 A | O | 1-339 | PDB |
| 7ZWD | EM | 300 A | O | 1-339 | PDB |
| 7ZX7 | EM | 340 A | O | 1-339 | PDB |
| 7ZX8 | EM | 300 A | O | 1-339 | PDB |
| 7ZXE | EM | 350 A | O | 1-339 | PDB |
| 8BVW | EM | 400 A | O | 1-339 | PDB |
| 8BYQ | EM | 410 A | O | 1-339 | PDB |
| 8BZ1 | EM | 380 A | O | 1-339 | PDB |
| 8GXQ | EM | 504 A | DP | 1-339 | PDB |
| 8GXS | EM | 416 A | DP | 1-339 | PDB |
| 8ITY | EM | 390 A | U | 1-339 | PDB |
| 8IUE | EM | 410 A | U | 1-339 | PDB |
| 8IUH | EM | 340 A | U | 1-339 | PDB |
| 8WAK | EM | 547 A | P | 1-339 | PDB |
| 8WAL | EM | 852 A | P | 1-339 | PDB |
| 8WAN | EM | 607 A | P | 1-339 | PDB |
| 8WAO | EM | 640 A | P | 1-339 | PDB |
| 8WAP | EM | 585 A | P | 1-339 | PDB |
| 8WAQ | EM | 629 A | P | 1-339 | PDB |
| 8WAR | EM | 720 A | P | 1-339 | PDB |
| 8WAS | EM | 613 A | P | 1-339 | PDB |
| AF-P20226-F1 | Predicted | AlphaFoldDB |
167 variants for P20226
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000625151 rs71815788 RCV001683617 |
95 | Q>missing | Spinocerebellar ataxia type 17 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs142540266 CA4108372 RCV001336551 |
137 | P>L | Spinocerebellar ataxia type 17 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs751702127 CA4108223 |
2 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA4108224 rs757450205 |
5 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs766191843 CA4108225 |
6 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA152221959 rs1009942407 |
8 | P>L | No |
ClinGen TOPMed |
|
|
CA4108227 rs764142412 |
11 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA152221976 rs949315152 |
17 | P>A | No |
ClinGen TOPMed |
|
|
rs1390001535 CA366516910 |
19 | G>V | No |
ClinGen gnomAD |
|
|
rs758309293 CA4108249 |
20 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA152223761 rs970666552 |
24 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA4108251 rs558863660 |
26 | P>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs150312586 CA4108253 |
27 | I>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs150312586 CA4108252 |
27 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4108255 rs577070091 |
31 | M>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs746303115 CA4108254 |
31 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs776116052 CA4108256 |
32 | M>R | No |
ClinGen ExAC |
|
|
CA366517007 rs1394976596 |
34 | Y>F | No |
ClinGen TOPMed |
|
|
CA4108258 rs749788903 |
35 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA366517032 rs1583127364 |
39 | T>P | No |
ClinGen Ensembl |
|
|
rs375575792 CA4108260 |
40 | P>A | No |
ClinGen ESP ExAC gnomAD |
|
|
CA4108261 rs761867883 |
44 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
CA4108262 rs772344160 |
47 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366517111 rs773389193 |
51 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773389193 CA4108263 |
51 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366517132 rs1190072930 |
54 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs764966099 CA4108269 |
58 | Q>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764966099 CA366517167 |
58 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752498284 CA4108273 |
60 | Q>H | No |
ClinGen ExAC TOPMed |
|
|
CA152223829 rs559386285 |
62 | Q>H | No |
ClinGen 1000Genomes TOPMed |
|
|
CA366517202 rs1215217094 |
63 | Q>* | No |
ClinGen gnomAD |
|
|
rs762804204 CA4108280 |
67 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1331989064 CA366517266 |
71 | Q>P | No |
ClinGen gnomAD |
|
|
rs55736770 CA152223968 |
72 | Q>H | No |
ClinGen TOPMed |
|
|
rs1410409146 CA366517279 |
73 | Q>E | No |
ClinGen gnomAD |
|
|
rs764035011 CA366517284 |
73 | Q>H | No |
ClinGen ExAC TOPMed |
|
|
CA366517285 rs764035011 |
73 | Q>H | No |
ClinGen ExAC TOPMed |
|
|
CA366517282 rs1229412131 |
73 | Q>R | No |
ClinGen TOPMed |
|
|
rs62430309 CA152223976 |
74 | Q>H | No |
ClinGen TOPMed |
|
|
rs56241301 CA152223998 |
75 | Q>H | No |
ClinGen ExAC TOPMed |
|
|
rs1197414368 CA366517298 |
75 | Q>L | No |
ClinGen TOPMed |
|
|
CA366517306 rs112083427 |
76 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1336548568 CA366517303 |
76 | Q>R | No |
ClinGen TOPMed |
|
|
RCV001692125 RCV000455402 rs1491487452 |
77 | Q>missing | No |
ClinVar dbSNP |
|
|
RCV001725920 rs771725566 RCV001725919 |
77 | Q>missing | No |
ClinVar dbSNP |
|
|
CA366517310 rs1346721481 |
77 | Q>* | No |
ClinGen gnomAD |
|
|
CA366517308 rs1346721481 |
77 | Q>K | No |
ClinGen gnomAD |
|
|
rs1198340310 CA366517318 |
78 | Q>* | No |
ClinGen gnomAD |
|
|
rs113440919 CA4108319 |
78 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1198340310 CA366517316 |
78 | Q>K | No |
ClinGen gnomAD |
|
|
CA4108320 rs780283741 |
79 | Q>R | No |
ClinGen ExAC |
|
|
CA366517332 rs1485539992 |
80 | Q>* | No |
ClinGen gnomAD |
|
|
CA366517333 rs1309337484 |
80 | Q>R | No |
ClinGen gnomAD |
|
|
rs779471026 CA366517345 |
81 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1471864851 CA366517346 |
82 | Q>* | No |
ClinGen gnomAD |
|
|
rs748266569 CA4108325 |
82 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366517354 rs1378238070 |
83 | Q>* | No |
ClinGen gnomAD |
|
|
CA366517373 rs1401305444 |
85 | Q>R | No |
ClinGen gnomAD |
|
|
CA366517379 rs1236899437 |
86 | Q>* | No |
ClinGen gnomAD |
|
|
CA366517390 rs1309553160 |
87 | Q>R | No |
ClinGen gnomAD |
|
|
CA366517398 rs1562359677 |
88 | Q>R | No |
ClinGen Ensembl |
|
|
CA4108333 rs764009292 |
89 | Q>R | No |
ClinGen ExAC TOPMed |
|
|
CA152224089 rs982117967 |
90 | Q>R | No |
ClinGen TOPMed |
|
|
CA152224114 rs796895015 |
92 | Q>H | No |
ClinGen TOPMed |
|
|
CA152224109 rs865926586 |
92 | Q>R | No |
ClinGen Ensembl |
|
|
rs1156940638 CA366517433 |
93 | Q>R | No |
ClinGen TOPMed |
|
|
RCV000736062 rs752404282 |
93 | Q>missing | No |
ClinVar dbSNP |
|
|
CA366517438 rs1273872785 |
94 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
rs143655307 CA4108344 |
95 | Q>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs752404282 RCV000736063 |
95 | Q>missing | No |
ClinVar dbSNP |
|
|
rs1043227165 CA152224130 |
96 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA366517479 rs1195703655 |
100 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs754217817 CA4108349 |
102 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754217817 CA4108348 |
102 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779443911 CA4108350 |
105 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs148074761 CA4108351 |
106 | T>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4108352 rs750797320 |
106 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs148074761 CA366517512 |
106 | T>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs148074761 CA366517513 |
106 | T>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4108354 rs747033709 |
108 | Q>R | No |
ClinGen ExAC TOPMed |
|
|
CA4108355 rs771116530 |
110 | A>T | No |
ClinGen ExAC |
|
|
rs201331220 CA152224166 |
111 | T>A | No |
ClinGen 1000Genomes gnomAD |
|
|
CA366517551 rs1399553263 |
112 | Q>P | No |
ClinGen TOPMed |
|
|
CA4108358 rs749081341 |
113 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs774242068 CA4108360 |
115 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366517577 rs1325318897 |
116 | G>V | No |
ClinGen Ensembl |
|
|
CA4108361 rs200427038 |
118 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4108363 rs772911295 |
119 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA4108362 rs767501232 |
119 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA152224194 rs908493900 |
120 | Q>E | No |
ClinGen Ensembl |
|
|
rs1562359838 CA366517604 |
121 | L>F | No |
ClinGen Ensembl |
|
|
rs1422934148 CA366517618 |
123 | H>Y | No |
ClinGen TOPMed |
|
|
CA366517645 rs1489716387 |
127 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs760224751 CA4108364 |
128 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs1208150616 CA366517650 |
128 | T>P | No |
ClinGen gnomAD |
|
|
CA4108365 rs760224751 |
128 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs1475609701 CA366517659 |
129 | T>I | No |
ClinGen TOPMed |
|
|
CA366517654 rs1465545382 |
129 | T>P | No |
ClinGen gnomAD |
|
|
rs941287680 CA152224204 |
131 | P>A | No |
ClinGen Ensembl |
|
|
rs753505824 CA4108366 |
131 | P>H | No |
ClinGen ExAC gnomAD |
|
|
rs199616061 CA4108367 |
133 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4108369 rs752995266 |
134 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4108370 rs140089853 |
136 | T>A | No |
ClinGen ESP ExAC TOPMed |
|
|
CA366517697 rs1370130375 |
136 | T>I | No |
ClinGen gnomAD |
|
|
rs778241571 CA4108371 |
137 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA152224228 rs778241571 |
137 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366517733 rs1583128175 |
143 | M>V | No |
ClinGen Ensembl |
|
|
CA4108376 rs769870393 |
146 | M>I | No |
ClinGen ExAC |
|
|
CA4108375 rs746127880 |
146 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA152224246 rs1028882699 |
147 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
rs747940424 CA4108378 |
150 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs773248432 CA4108380 |
152 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1329917969 CA366517792 |
152 | A>V | No |
ClinGen gnomAD |
|
|
CA366517799 rs1218192424 |
153 | T>M | No |
ClinGen TOPMed |
|
|
CA366517864 rs1422030633 |
163 | P>L | No |
ClinGen gnomAD |
|
|
CA366517873 rs1412145625 |
165 | L>M | No |
ClinGen gnomAD |
|
|
CA366518171 rs1312590130 |
170 | S>Y | No |
ClinGen gnomAD |
|
|
CA366518232 rs1412175500 |
175 | G>C | No |
ClinGen gnomAD |
|
|
CA366518237 rs1367954958 |
175 | G>V | No |
ClinGen Ensembl |
|
|
CA4108407 rs375063272 |
176 | C>R | No |
ClinGen ESP ExAC gnomAD |
|
|
CA152225202 rs112100346 |
182 | T>A | No |
ClinGen Ensembl |
|
|
CA4108408 rs762389131 |
182 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA366518368 rs1299337339 |
184 | A>G | No |
ClinGen TOPMed |
|
|
COSM1076233 rs756227958 CA4108411 |
186 | R>C | endometrium haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed |
|
CA366518415 rs1209911056 |
188 | R>* | No |
ClinGen gnomAD |
|
|
rs1419445585 CA366518512 |
200 | V>I | No |
ClinGen gnomAD |
|
|
rs1419445585 CA366518514 |
200 | V>L | No |
ClinGen gnomAD |
|
|
rs1406414133 CA366518541 |
204 | I>L | No |
ClinGen TOPMed gnomAD |
|
|
CA4108442 rs745830790 |
206 | E>A | No |
ClinGen ExAC |
|
|
rs756008941 CA4108443 |
208 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs1583131728 CA366518572 |
209 | T>P | No |
ClinGen Ensembl |
|
|
rs748740885 CA4108445 |
210 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs1583131737 CA366518600 |
213 | I>M | No |
ClinGen Ensembl |
|
|
rs887521451 CA152226094 |
215 | S>T | No |
ClinGen Ensembl |
|
|
rs1295992676 CA366518621 |
216 | S>C | No |
ClinGen gnomAD |
|
|
rs748590964 CA366518640 |
219 | M>R | No |
ClinGen ExAC gnomAD |
|
|
rs748590964 CA4108448 |
219 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA366518648 rs1431326572 |
220 | V>A | No |
ClinGen TOPMed |
|
|
rs1269326640 CA366518659 |
222 | T>A | No |
ClinGen gnomAD |
|
|
rs1188907199 CA366518680 |
225 | K>R | No |
ClinGen TOPMed |
|
|
rs866919321 CA152227236 |
229 | Q>K | No |
ClinGen Ensembl |
|
|
CA4108470 rs747363929 |
230 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA366518809 rs1333457265 |
242 | Q>* | No |
ClinGen gnomAD |
|
|
CA366518885 rs759710446 |
252 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA4108473 rs759710446 |
252 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs1246604155 CA366518917 |
257 | N>H | No |
ClinGen TOPMed |
|
|
CA4108474 rs775603111 |
258 | M>V | No |
ClinGen ExAC |
|
|
CA366518998 rs1206466089 |
268 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1018596766 CA152227283 |
271 | E>D | No |
ClinGen TOPMed |
|
|
rs1008548886 CA152227280 |
271 | E>G | No |
ClinGen TOPMed |
|
|
CA366519030 rs1305796742 |
273 | L>V | No |
ClinGen TOPMed |
|
|
CA366519046 rs1365457491 |
275 | L>P | No |
ClinGen TOPMed |
|
|
rs1298121577 CA366519208 |
296 | I>L | No |
ClinGen gnomAD |
|
|
CA366519227 rs1325232464 |
298 | P>L | No |
ClinGen gnomAD |
|
|
CA366519238 rs1562363181 |
300 | I>F | No |
ClinGen Ensembl |
|
|
CA4108496 rs774531109 |
305 | F>S | No |
ClinGen ExAC gnomAD |
|
|
rs774673727 CA4108497 |
306 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs533679967 CA4108498 |
308 | G>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1420558024 CA366519349 |
316 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
rs904248845 CA366519355 |
316 | K>N | No |
ClinGen gnomAD |
|
|
rs1420558024 CA366519348 |
316 | K>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1379121744 CA366519381 |
320 | E>D | No |
ClinGen gnomAD |
|
|
rs762934477 CA4108525 |
328 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA366519595 rs1349833584 |
336 | R>K | No |
ClinGen TOPMed |
|
|
rs1347718096 CA366519602 |
336 | R>S | No |
ClinGen gnomAD |
|
|
rs751047986 CA4108527 COSM1162817 |
338 | T>M | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA4108529 rs372970346 |
339 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
1 associated diseases with P20226
[MIM: 607136]: Spinocerebellar ataxia 17 (SCA17)
Spinocerebellar ataxia is a clinically and genetically heterogeneous group of cerebellar disorders. Patients show progressive incoordination of gait and often poor coordination of hands, speech and eye movements, due to degeneration of the cerebellum with variable involvement of the brainstem and spinal cord. SCA17 is an autosomal dominant cerebellar ataxia (ADCA) characterized by widespread cerebral and cerebellar atrophy, dementia and extrapyramidal signs. The molecular defect in SCA17 is the expansion of a CAG repeat in the coding region of TBP. Longer expansions result in earlier onset and more severe clinical manifestations of the disease. {ECO:0000269|PubMed:11313753, ECO:0000269|PubMed:11448935, ECO:0000269|PubMed:11939898}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- Spinocerebellar ataxia is a clinically and genetically heterogeneous group of cerebellar disorders. Patients show progressive incoordination of gait and often poor coordination of hands, speech and eye movements, due to degeneration of the cerebellum with variable involvement of the brainstem and spinal cord. SCA17 is an autosomal dominant cerebellar ataxia (ADCA) characterized by widespread cerebral and cerebellar atrophy, dementia and extrapyramidal signs. The molecular defect in SCA17 is the expansion of a CAG repeat in the coding region of TBP. Longer expansions result in earlier onset and more severe clinical manifestations of the disease. {ECO:0000269|PubMed:11313753, ECO:0000269|PubMed:11448935, ECO:0000269|PubMed:11939898}. Note=The disease is caused by variants affecting the gene represented in this entry.
1 regional properties for P20226
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | TATA-box binding protein, conserved site | 283 - 332 | IPR030491 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR10126 | TATA-BOX BINDING PROTEIN |
| PANTHER Subfamily | PTHR10126:SF20 | TATA-BOX-BINDING PROTEIN |
| PANTHER Protein Class |
RNA metabolism protein
general transcription factor |
|
| PANTHER Pathway Category |
Huntington disease TBP General transcription by RNA polymerase I TBP Transcription regulation by bZIP transcription factor TBP |
|
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| euchromatin | A dispersed and relatively uncompacted form of chromatin that is in a transcription-competent conformation. |
| female pronucleus | The pronucleus originating from the ovum that is being fertilized. |
| male pronucleus | The pronucleus originating from the spermatozoa that was involved in fertilization. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| transcription factor TFIIA complex | A component of the transcription machinery of RNA Polymerase II. In humans, TFIIA is a heterotrimer composed of an alpha (P35), beta (P19) and gamma subunits (P12). |
| transcription factor TFIID complex | A complex composed of TATA binding protein (TBP) and TBP associated factors (TAFs); the total mass is typically about 800 kDa. Most of the TAFs are conserved across species. In TATA-containing promoters for RNA polymerase II (Pol II), TFIID is believed to recognize at least two distinct elements, the TATA element and a downstream promoter element. TFIID is also involved in recognition of TATA-less Pol II promoters. Binding of TFIID to DNA is necessary but not sufficient for transcription initiation from most RNA polymerase II promoters. |
| transcription preinitiation complex | A protein-DNA complex composed of proteins binding promoter DNA to form the transcriptional preinitiation complex (PIC), the formation of which is a prerequisite for transcription. |
12 GO annotations of molecular function
| Name | Definition |
|---|---|
| aryl hydrocarbon receptor binding | Binding to an aryl hydrocarbon receptor. |
| core promoter sequence-specific DNA binding | Binding to a sequence of DNA that is part of a core promoter region. The core promoter is composed of the transcription start site and binding sites for the RNA polymerase and the basal transcription machinery. The transcribed region might be described as a gene, cistron, or operon. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| general transcription initiation factor activity | A molecular function required for core promoter activity that mediates the assembly of the RNA polymerase holoenzyme at promoter DNA to form the pre-initiation complex (PIC). General transcription factors (GTFs) bind to and open promoter DNA, initiate RNA synthesis and stimulate the escape of the polymerase from the promoter. Not all subunits of the general transcription factor are necessarily present at all promoters to initiate transcription. GTFs act at each promoter, although the exact subunit composition at individual promoters may vary. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| RNA polymerase II core promoter sequence-specific DNA binding | Binding to a DNA sequence that is part of the core promoter of a RNA polymerase II-transcribed gene. |
| RNA polymerase II general transcription initiation factor activity | A general transcription initiation factor activity that contributes to transcription start site selection and transcription initiation of genes transcribed by RNA polymerase II. The general transcription factors for RNA polymerase II include TFIIB, TFIID, TFIIE, TFIIF, TFIIH and TATA-binding protein (TBP). In most species, RNA polymerase II transcribes all messenger RNAs (mRNAs), most untranslated regulatory RNAs, the majority of the snoRNAs, four of the five snRNAs (U1, U2, U4, and U5), and other small noncoding RNAs. For some small RNAs there is variability between species as to whether it is transcribed by RNA polymerase II or RNA polymerase III. However there are also rare exceptions, such as Trypanosoma brucei, where RNA polymerase I transcribes certain mRNAs in addition to its normal role in rRNA transcription. |
| RNA polymerase II general transcription initiation factor binding | Binding to a basal RNA polymerase II transcription factor, any of the factors involved in formation of the preinitiation complex (PIC) by RNA polymerase II and defined as a basal or general transcription factor. |
| RNA polymerase III general transcription initiation factor activity | A general transcription initiation factor activity that contributes to transcription start site selection and transcription initiation of genes transcribed by RNA polymerase III. Factors required for RNA polymerase III transcription initiation include TFIIIA, TFIIIB and TFIIIC. RNA polymerase III transcribes genes encoding short RNAs, including tRNAs, 5S rRNA, U6 snRNA, the short ncRNA component of RNases P, the mitochondrial RNA processing (MRP) RNA, the signal recognition particle SRP RNA, and in higher eukaryotes a number of micro and other small RNAs, though there is some variability across species as to whether a given small noncoding RNA is transcribed by RNA polymerase II or RNA polymerase III. |
| TFIIB-class transcription factor binding | Binding to a general RNA polymerase II transcription factor of the TFIIB class, one of the factors involved in formation of the preinitiation complex (PIC) by RNA polymerase II. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| DNA-templated transcription, initiation | The initial step of transcription, consisting of the assembly of the RNA polymerase preinitiation complex (PIC) at a gene promoter, as well as the formation of the first few bonds of the RNA transcript. Transcription initiation includes abortive initiation events, which occur when the first few nucleotides are repeatedly synthesized and then released, and ends when promoter clearance takes place. |
| mRNA transcription by RNA polymerase II | The cellular synthesis of messenger RNA (mRNA) from a DNA template by RNA polymerase II, originating at an RNA polymerase II promoter. |
| positive regulation of transcription initiation from RNA polymerase II promoter | Any process that increases the rate, frequency or extent of a process involved in starting transcription from an RNA polymerase II promoter. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| RNA polymerase II preinitiation complex assembly | The aggregation, arrangement and bonding together of proteins on an RNA polymerase II promoter DNA to form the transcriptional preinitiation complex (PIC), the formation of which is a prerequisite for transcription by RNA polymerase. |
| transcription by RNA polymerase II | The synthesis of RNA from a DNA template by RNA polymerase II (RNAP II), originating at an RNA polymerase II promoter. Includes transcription of messenger RNA (mRNA) and certain small nuclear RNAs (snRNAs). |
| transcription by RNA polymerase III | The synthesis of RNA from a DNA template by RNA polymerase III, originating at an RNAP III promoter. |
| transcription initiation from RNA polymerase II promoter | A transcription initiation process that takes place at a RNA polymerase II gene promoter. Messenger RNAs (mRNA) genes, as well as some non-coding RNAs, are transcribed by RNA polymerase II. |
24 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P26356 | TBP1 | TATA-box-binding protein 1 | Triticum aestivum (Wheat) | PR |
| Q02879 | TBP2 | TATA-box-binding protein 2 | Triticum aestivum (Wheat) | PR |
| P13393 | SPT15 | TATA-box-binding protein | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | EV |
| Q2HJ52 | TBP | TATA-box-binding protein | Bos taurus (Bovine) | PR |
| O13270 | TBP | TATA-box-binding protein | Gallus gallus (Chicken) | PR |
| A6H907 | TBPL2 | TATA box-binding protein-like 2 | Pan troglodytes (Chimpanzee) | PR |
| Q27896 | Trf | TBP-related factor | Drosophila melanogaster (Fruit fly) | PR |
| Q6SJ96 | TBPL2 | TATA box-binding protein-like 2 | Homo sapiens (Human) | PR |
| P50159 | TBP2 | TATA-box-binding protein 2 | Zea mays (Maize) | PR |
| P50158 | TBP1 | TATA-box-binding protein 1 | Zea mays (Maize) | PR |
| Q6SJ95 | Tbpl2 | TATA box-binding protein-like 2 | Mus musculus (Mouse) | PR |
| P29037 | Tbp | TATA-box-binding protein | Mus musculus (Mouse) | PR |
| P26357 | TBP | TATA-box-binding protein | Solanum tuberosum (Potato) | PR |
| A6H909 | Tbpl2 | TATA box-binding protein-like 2 | Rattus norvegicus (Rat) | PR |
| Q8W0W4 | TBP2 | TATA-binding protein 2 | Oryza sativa subsp japonica (Rice) | PR |
| B2D6P4 | tlf-1 | TATA box-binding protein-like 1 | Caenorhabditis elegans | SS |
| O45211 | Tbp | TATA-box-binding protein | Bombyx mori (Silk moth) | PR |
| Q42808 | TBP1 | TATA-box-binding protein | Glycine max (Soybean) (Glycine hispida) | PR |
| P28148 | TBP2 | TATA-box-binding protein 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P28147 | TBP1 | TATA-box-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q28DH2 | tbpl2 | TATA box-binding protein-like 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q28GG8 | tbp | TATA-box-binding protein | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q1JPY4 | tbpl2 | TATA box-binding protein-like 2 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q7SXL3 | tbp | TATA-box-binding protein | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDQNNSLPPY | AQGLASPQGA | MTPGIPIFSP | MMPYGTGLTP | QPIQNTNSLS | ILEEQQRQQQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QQQQQQQQQQ | QQQQQQQQQQ | QQQQQQQQQQ | QQQQQAVAAA | AVQQSTSQQA | TQGTSGQAPQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LFHSQTLTTA | PLPGTTPLYP | SPMTPMTPIT | PATPASESSG | IVPQLQNIVS | TVNLGCKLDL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KTIALRARNA | EYNPKRFAAV | IMRIREPRTT | ALIFSSGKMV | CTGAKSEEQS | RLAARKYARV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VQKLGFPAKF | LDFKIQNMVG | SCDVKFPIRL | EGLVLTHQQF | SSYEPELFPG | LIYRMIKPRI |
| 310 | 320 | 330 | |||
| VLLIFVSGKV | VLTGAKVRAE | IYEAFENIYP | ILKGFRKTT |