P18654
Gene name |
Rps6ka3 |
Protein name |
Ribosomal protein S6 kinase alpha-3 |
Names |
S6K-alpha-3, 90 kDa ribosomal protein S6 kinase 3, p90-RSK 3, p90RSK3, MAP kinase-activated protein kinase 1b, MAPK-activated protein kinase 1b, MAPKAP kinase 1b, MAPKAPK-1b, Ribosomal S6 kinase 2, RSK-2, pp90RSK2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:110651 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
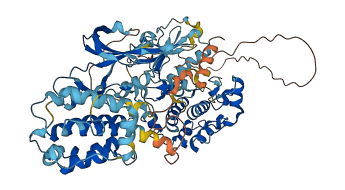
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
422-679 (Protein kinase domain) |
Relief mechanism |
PTM, Partner binding |
Assay |
|
Accessory elements
210-233 (Activation loop from InterPro)
Target domain |
72-388 (N-terminal catalytic domain of the Serine/Threonine Kinase, 90 kDa ribosomal protein S6 kinase) |
Relief mechanism |
|
Assay |
|
560-583 (Activation loop from InterPro)
Target domain |
402-740 (C-terminal catalytic domain of the Serine/Threonine Kinase, Ribosomal S6 kinase 2, also called 90kDa ribosomal protein S6 kinase 3 or Ribosomal protein S6 kinase alpha-3) |
Relief mechanism |
|
Assay |
|
References
- Li D et al. (2012) "Structural basis for the autoinhibition of the C-terminal kinase domain of human RSK1", Acta crystallographica. Section D, Biological crystallography, 68, 680-5
- Li D et al. (2013) "The prometastatic ribosomal S6 kinase 2-cAMP response element-binding protein (RSK2-CREB) signaling pathway up-regulates the actin-binding protein fascin-1 to promote tumor metastasis", The Journal of biological chemistry, 288, 32528-32538
- Malakhova M et al. (2008) "Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2", Nature structural & molecular biology, 15, 112-3
- Poteet-Smith CE et al. (1999) "Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family", The Journal of biological chemistry, 274, 22135-8
Autoinhibited structure

Activated structure

10 structures for P18654
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2QR7 | X-ray | 200 A | A | 399-740 | PDB |
| 2QR8 | X-ray | 200 A | A | 399-740 | PDB |
| 3G51 | X-ray | 180 A | A | 44-367 | PDB |
| 3UBD | X-ray | 153 A | A | 45-346 | PDB |
| 4EL9 | X-ray | 155 A | A | 45-346 | PDB |
| 4GUE | X-ray | 180 A | A | 45-346 | PDB |
| 4M8T | X-ray | 300 A | A | 399-740 | PDB |
| 4MAO | X-ray | 260 A | A | 399-740 | PDB |
| 5O1S | X-ray | 190 A | A | 400-740 | PDB |
| AF-P18654-F1 | Predicted | AlphaFoldDB |
No variants for P18654
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P18654 | |||||
No associated diseases with P18654
10 regional properties for P18654
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 68 - 327 | IPR000719-1 |
| domain | Protein kinase domain | 422 - 679 | IPR000719-2 |
| domain | AGC-kinase, C-terminal | 328 - 397 | IPR000961 |
| active_site | Serine/threonine-protein kinase, active site | 189 - 201 | IPR008271-1 |
| active_site | Serine/threonine-protein kinase, active site | 535 - 547 | IPR008271-2 |
| binding_site | Protein kinase, ATP binding site | 74 - 100 | IPR017441-1 |
| binding_site | Protein kinase, ATP binding site | 428 - 451 | IPR017441-2 |
| domain | Protein kinase, C-terminal | 351 - 387 | IPR017892 |
| domain | Ribosomal protein S6 kinase alpha-3, C-terminal catalytic domain | 402 - 740 | IPR041905 |
| domain | Ribosomal S6 kinase, N-terminal catalytic domain | 72 - 388 | IPR041906 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| cysteine-type endopeptidase inhibitor activity involved in apoptotic process | Binds to and stops, prevents or reduces the activity of a cysteine-type endopeptidase involved in the apoptotic process. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| ribosomal protein S6 kinase activity | Catalysis of the reaction: ribosomal protein S6 + ATP = ribosomal protein S6 phosphate + ATP. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of a cysteine-type endopeptidase activity involved in the apoptotic process. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| toll-like receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a toll-like receptor on the surface of a target cell. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
19 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P18652 | RPS6KA | Ribosomal protein S6 kinase 2 alpha | Gallus gallus (Chicken) | SS |
| Q5F3L1 | RPS6KA5 | Ribosomal protein S6 kinase alpha-5 | Gallus gallus (Chicken) | SS |
| Q9UK32 | RPS6KA6 | Ribosomal protein S6 kinase alpha-6 | Homo sapiens (Human) | SS |
| O75676 | RPS6KA4 | Ribosomal protein S6 kinase alpha-4 | Homo sapiens (Human) | SS |
| Q96S38 | RPS6KC1 | Ribosomal protein S6 kinase delta-1 | Homo sapiens (Human) | PR |
| Q15349 | RPS6KA2 | Ribosomal protein S6 kinase alpha-2 | Homo sapiens (Human) | SS |
| Q15418 | RPS6KA1 | Ribosomal protein S6 kinase alpha-1 | Homo sapiens (Human) | EV |
| O75582 | RPS6KA5 | Ribosomal protein S6 kinase alpha-5 | Homo sapiens (Human) | SS |
| P51812 | RPS6KA3 | Ribosomal protein S6 kinase alpha-3 | Homo sapiens (Human) | EV |
| Q8BLK9 | Rps6kc1 | Ribosomal protein S6 kinase delta-1 | Mus musculus (Mouse) | PR |
| P18653 | Rps6ka1 | Ribosomal protein S6 kinase alpha-1 | Mus musculus (Mouse) | SS |
| Q9WUT3 | Rps6ka2 | Ribosomal protein S6 kinase alpha-2 | Mus musculus (Mouse) | SS |
| Q9Z2B9 | Rps6ka4 | Ribosomal protein S6 kinase alpha-4 | Mus musculus (Mouse) | SS |
| Q8C050 | Rps6ka5 | Ribosomal protein S6 kinase alpha-5 | Mus musculus (Mouse) | PR |
| Q9Z1M4 | Rps6kb2 | Ribosomal protein S6 kinase beta-2 | Mus musculus (Mouse) | SS |
| Q8BSK8 | Rps6kb1 | Ribosomal protein S6 kinase beta-1 | Mus musculus (Mouse) | SS |
| Q63531 | Rps6ka1 | Ribosomal protein S6 kinase alpha-1 | Rattus norvegicus (Rat) | SS |
| Q18846 | rskn-2 | Putative ribosomal protein S6 kinase alpha-2 | Caenorhabditis elegans | PR |
| Q21734 | rskn-1 | Putative ribosomal protein S6 kinase alpha-1 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPLAQLADPW | QKMAVESPSD | SAENGQQIMD | EPMGEEEINP | QTEEGSIKEI | AITHHVKEGH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EKADPSQFEL | LKVLGQGSFG | KVFLVKKISG | SDARQLYAMK | VLKKATLKVR | DRVRTKMERD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ILVEVNHPFI | VKLHYAFQTE | GKLYLILDFL | RGGDLFTRLS | KEVMFTEEDV | KFYLAELALA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LDHLHSLGII | YRDLKPENIL | LDEEGHIKLT | DFGLSKESID | HEKKAYSFCG | TVEYMAPEVV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NRRGHTQSAD | WWSFGVLMFE | MLTGTLPFQG | KDRKETMTMI | LKAKLGMPQF | LSPEAQSLLR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MLFKRNPANR | LGAGPDGVEE | IKRHSFFSTI | DWNKLYRREI | HPPFKPATGR | PEDTFYFDPE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FTAKTPKDSP | GIPPSANAHQ | LFRGFSFVAI | TSDDESQAMQ | TVGVHSIVQQ | LHRNSIQFTD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GYEVKEDIGV | GSYSVCKRCI | HKATNMEFAV | KIIDKSKRDP | TEEIEILLRY | GQHPNIITLK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DVYDDGKYVY | VVTELMKGGE | LLDKILRQKF | FSEREASAVL | FTITKTVEYL | HAQGVVHRDL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| KPSNILYVDE | SGNPESIRIC | DFGFAKQLRA | ENGLLMTPCY | TANFVAPEVL | KRQGYDAACD |
| 610 | 620 | 630 | 640 | 650 | 660 |
| IWSLGVLLYT | MLTGYTPFAN | GPDDTPEEIL | ARIGSGKFSL | SGGYWNSVSD | TAKDLVSKML |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HVDPHQRLTA | ALVLRHPWIV | HWDQLPQYQL | NRQDAPHLVK | GAMAATYSAL | NRNQSPVLEP |
| 730 | |||||
| VGRSTLAQRR | GIKKITSTAL |