P18085
Gene name |
ARF4 (ARF2) |
Protein name |
ADP-ribosylation factor 4 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:378 |
EC number |
|
Protein Class |
ADP RIBOSYLATION FACTOR-RELATED (PTHR11711) |
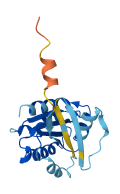
Descriptions
Small GTPases couple their GDP/GTP structural cycle to cytosol/membrane alternation to function as versatile molecular switches in the cell. Membrane localization of their active, GTP-bound form is pivotal to their ability to propagate information, and this requires their post-translational modification by lipids.
Arf GTPases are modified by a myristate attached to their N-terminus, which is shielded by intramolecular interactions in their inactive state. The myristoylated N-terminus of Arf is autoinhibitory in solution and is displaced by membranes, priming Arf GTPases for activation by their GEFs. Replacement of the N-terminal myristate by a 6xHis-tag preserves autoinhibition, representing that membranes unlock the N-terminal region to facilitate subsequent activation.
Autoinhibitory domains (AIDs)
Target domain |
16-140 (Small GTP-binding protein domain) |
Relief mechanism |
Ligand binding |
Assay |
Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for P18085
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1Z6X | X-ray | 270 A | A/B | 1-180 | PDB |
| AF-P18085-F1 | Predicted | AlphaFoldDB |
97 variants for P18085
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| COSM1424901 | 1 | M>? | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1316437277 | 4 | T>I | No |
TOPMed gnomAD |
|
| rs1447051765 | 5 | I>T | No | gnomAD | |
| rs769987303 | 5 | I>V | No |
ExAC gnomAD |
|
| rs2070199209 | 8 | L>F | No | TOPMed | |
| rs922610512 | 10 | S>F | No | TOPMed | |
| rs1317538168 | 11 | R>* | No | gnomAD | |
| rs1224376043 | 11 | R>Q | No | gnomAD | |
| rs777601584 | 14 | G>R | No |
ExAC gnomAD |
|
| rs777601584 | 14 | G>S | No |
ExAC gnomAD |
|
| rs1221145627 | 14 | G>V | No | gnomAD | |
| rs2070198866 | 15 | K>R | No | TOPMed | |
| rs772174333 | 20 | I>L | No |
ExAC TOPMed gnomAD |
|
| COSM4119552 | 22 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1161084466 | 23 | V>A | No | gnomAD | |
| rs1327085319 | 23 | V>I | No |
TOPMed gnomAD |
|
| rs748165241 | 24 | G>R | No |
ExAC gnomAD |
|
| rs1385837777 | 26 | D>G | No | gnomAD | |
| COSM3596712 | 26 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2153408996 | 31 | T>I | No | Ensembl | |
| rs768064782 | 34 | L>R | No |
ExAC gnomAD |
|
| rs1485460096 | 39 | L>V | No | Ensembl | |
| rs748926137 | 41 | E>V | No |
ExAC gnomAD |
|
| rs779606925 | 42 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs2070012656 | 43 | V>I | No | gnomAD | |
| rs755833114 | 46 | I>V | No |
ExAC gnomAD |
|
|
TCGA novel rs1575784499 |
50 | G>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| COSM3596711 | 52 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs867291321 | 55 | T>A | No | Ensembl | |
| rs1281054796 | 56 | V>A | No | gnomAD | |
| rs772444015 | 57 | E>D | No |
ExAC gnomAD |
|
| rs1406321497 | 57 | E>K | No | gnomAD | |
| rs748613969 | 59 | K>E | No |
ExAC gnomAD |
|
| rs2070006405 | 60 | N>H | No | Ensembl | |
|
rs2070006325 COSM98335 |
61 | I>V | upper_aerodigestive_tract Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs2070006252 | 64 | T>I | No | Ensembl | |
|
rs11550597 VAR_048317 |
68 | V>A | No |
UniProt Ensembl dbSNP |
|
| TCGA novel | 73 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1413759346 | 73 | R>T | No | gnomAD | |
| rs1212221932 | 74 | I>V | No | Ensembl | |
| rs2070005963 | 80 | H>N | No | TOPMed | |
| rs1174556160 | 84 | N>S | No | gnomAD | |
| rs1432985527 | 85 | T>N | No |
TOPMed gnomAD |
|
| rs1283496925 | 88 | L>P | No |
TOPMed gnomAD |
|
| rs1409029602 | 88 | L>V | No |
TOPMed gnomAD |
|
| rs1575781288 | 89 | I>T | No | Ensembl | |
| rs1347064982 | 91 | V>M | No |
TOPMed gnomAD |
|
| TCGA novel | 94 | S>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1424900 | 95 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs367913501 | 95 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs2069918493 | 96 | D>N | No | gnomAD | |
| rs748489377 | 97 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs748489377 | 97 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs2069918463 | 97 | R>S | No | TOPMed | |
| rs938032119 | 98 | E>G | No | gnomAD | |
| COSM1047705 | 99 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4818693 | 99 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1245184248 | 100 | I>F | No |
TOPMed gnomAD |
|
| rs2069918261 | 102 | E>K | No | Ensembl | |
| rs1380540189 | 103 | V>A | No | gnomAD | |
| rs760385965 | 113 | V>I | No |
ExAC gnomAD |
|
| rs774513747 | 119 | A>T | No |
ExAC gnomAD |
|
| rs1166933486 | 125 | A>V | No |
TOPMed gnomAD |
|
| TCGA novel | 126 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2069894050 | 127 | K>R | No | TOPMed | |
| TCGA novel | 129 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 133 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749709237 | 135 | A>S | No |
ExAC gnomAD |
|
| rs749709237 | 135 | A>T | No |
ExAC gnomAD |
|
| rs1396134797 | 135 | A>V | No | Ensembl | |
| rs776097507 | 136 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1296191890 | 138 | E>D | No | gnomAD | |
| rs2069893692 | 140 | T>P | No | Ensembl | |
| rs770467489 | 141 | D>N | No |
ExAC gnomAD |
|
| rs1349818124 | 143 | L>V | No | gnomAD | |
| rs1258860922 | 144 | G>R | No | gnomAD | |
| rs781482985 | 146 | Q>H | No |
ExAC gnomAD |
|
| rs1559782595 | 146 | Q>R | No | Ensembl | |
|
COSM167927 rs747265204 |
149 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM192729 rs777379437 |
149 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs777379437 | 149 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs747265204 | 149 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs2069893249 | 151 | R>G | No | TOPMed | |
| rs2069893216 | 151 | R>T | No | TOPMed | |
| rs2069850932 | 155 | V>L | No | Ensembl | |
| rs1575778588 | 156 | Q>K | No | Ensembl | |
| rs1259731167 | 157 | A>D | No | TOPMed | |
| rs1468535094 | 164 | T>A | No | gnomAD | |
| rs2069850570 | 167 | Y>F | No | Ensembl | |
| rs1172787098 | 170 | L>F | No | gnomAD | |
| COSM1047704 | 171 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs80056234 | 174 | S>A | No | gnomAD | |
| rs2153408211 | 175 | N>D | No | Ensembl | |
|
rs778744712 COSM1692892 |
180 | R>C | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs754842478 | 180 | R>H | No |
ExAC gnomAD |
|
| rs1559781566 | 181 | R>Q | No | Ensembl | |
| rs138912511 | 181 | R>S | No |
ESP TOPMed |
No associated diseases with P18085
7 regional properties for P18085
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | CARD domain | 439 - 529 | IPR001315 |
| repeat | BIR repeat | 27 - 98 | IPR001370-1 |
| repeat | BIR repeat | 167 - 237 | IPR001370-2 |
| repeat | BIR repeat | 253 - 324 | IPR001370-3 |
| domain | Zinc finger, RING-type | 557 - 592 | IPR001841 |
| domain | BIRC2/BIRC3, UBA domain | 377 - 421 | IPR041933 |
| domain | BIRC2/3-like, UBA domain | 374 - 426 | IPR048875 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11711 | ADP RIBOSYLATION FACTOR-RELATED |
| PANTHER Subfamily | PTHR11711:SF110 | ADP-RIBOSYLATION FACTOR 4 |
| PANTHER Protein Class | G-protein | |
| PANTHER Pathway Category |
Huntington disease ARF |
|
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| epidermal growth factor receptor binding | Binding to an epidermal growth factor receptor. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
| guanyl-nucleotide exchange factor activity | Stimulates the exchange of GDP to GTP on a signaling GTPase, changing its conformation to its active form. Guanine nucleotide exchange factors (GEFs) act by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP), which is more abundant in the cell under normal cellular physiological conditions. |
| NAD+-protein-arginine ADP-ribosyltransferase activity | Catalysis of the reaction |
| phospholipase D activator activity | Binds to and increases the activity of the enzyme phospholipase D. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| apical protein localization | Any process in which a protein is transported to, or maintained in, apical regions of the cell. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. |
| cilium assembly | The assembly of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole. |
| dendritic spine development | The process whose specific outcome is the progression of the dendritic spine over time, from its formation to the mature structure. A dendritic spine is a protrusion from a dendrite and a specialized subcellular compartment involved in synaptic transmission. |
| endoplasmic reticulum to Golgi vesicle-mediated transport | The directed movement of substances from the endoplasmic reticulum (ER) to the Golgi, mediated by COP II vesicles. Small COP II coated vesicles form from the ER and then fuse directly with the cis-Golgi. Larger structures are transported along microtubules to the cis-Golgi. |
| establishment or maintenance of epithelial cell apical/basal polarity | Any cellular process that results in the specification, formation or maintenance of the apicobasal polarity of an epithelial cell. |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| learning | Any process in an organism in which a relatively long-lasting adaptive behavioral change occurs as the result of experience. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| protein localization to cilium | A process in which a protein is transported to, or maintained in, a location within a cilium. |
| regulation of cilium assembly | Any process that modulates the frequency, rate or extent of cilium assembly. |
| regulation of postsynapse organization | Any process that modulates the physical form of a postsynapse. |
| regulation of reactive oxygen species metabolic process | Any process that modulates the frequency, rate or extent of reactive oxygen species metabolic process. |
| retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | The directed movement of substances from the Golgi back to the endoplasmic reticulum, mediated by vesicles bearing specific protein coats such as COPI or COG. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
48 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJ96 | ARL5A | ADP-ribosylation factor-like protein 5A | Bos taurus (Bovine) | SS |
| P84081 | ARF2 | ADP-ribosylation factor 2 | Bos taurus (Bovine) | SS |
| P84080 | ARF1 | ADP-ribosylation factor 1 | Bos taurus (Bovine) | SS |
| Q0VC18 | ARL4D | ADP-ribosylation factor-like protein 4D | Bos taurus (Bovine) | PR |
| Q3SZF2 | ARF4 | ADP-ribosylation factor 4 | Bos taurus (Bovine) | SS |
| P49702 | ARF5 | ADP-ribosylation factor 5 | Gallus gallus (Chicken) | SS |
| P61209 | Arf1 | ADP-ribosylation factor 1 | Drosophila melanogaster (Fruit fly) | SS |
| P40945 | Arf4 | ADP ribosylation factor 4 | Drosophila melanogaster (Fruit fly) | SS |
| A6NH57 | ARL5C | Putative ADP-ribosylation factor-like protein 5C | Homo sapiens (Human) | SS |
| P49703 | ARL4D | ADP-ribosylation factor-like protein 4D | Homo sapiens (Human) | PR |
| P84077 | ARF1 | ADP-ribosylation factor 1 | Homo sapiens (Human) | EV |
| P84085 | ARF5 | ADP-ribosylation factor 5 | Homo sapiens (Human) | EV |
| Q8N4G2 | ARL14 | ADP-ribosylation factor-like protein 14 | Homo sapiens (Human) | SS |
| Q969Q4 | ARL11 | ADP-ribosylation factor-like protein 11 | Homo sapiens (Human) | PR |
| Q96KC2 | ARL5B | ADP-ribosylation factor-like protein 5B | Homo sapiens (Human) | SS |
| P62330 | ARF6 | ADP-ribosylation factor 6 | Homo sapiens (Human) | EV |
| Q8IVW1 | ARL17A | ADP-ribosylation factor-like protein 17 | Homo sapiens (Human) | SS |
| Q9H0F7 | ARL6 | ADP-ribosylation factor-like protein 6 | Homo sapiens (Human) | SS |
| P40616 | ARL1 | ADP-ribosylation factor-like protein 1 | Homo sapiens (Human) | SS |
| P61204 | ARF3 | ADP-ribosylation factor 3 | Homo sapiens (Human) | EV |
| Q9Y689 | ARL5A | ADP-ribosylation factor-like protein 5A | Homo sapiens (Human) | SS |
| P49076 | ARF1 | ADP-ribosylation factor | Zea mays (Maize) | SS |
| P84084 | Arf5 | ADP-ribosylation factor 5 | Mus musculus (Mouse) | SS |
| Q99PE9 | Arl4d | ADP-ribosylation factor-like protein 4D | Mus musculus (Mouse) | PR |
| Q6P068 | Arl5c | ADP-ribosylation factor-like protein 5C | Mus musculus (Mouse) | SS |
| Q80ZU0 | Arl5a | ADP-ribosylation factor-like protein 5A | Mus musculus (Mouse) | SS |
| P61205 | Arf3 | ADP-ribosylation factor 3 | Mus musculus (Mouse) | SS |
| Q9D4P0 | Arl5b | ADP-ribosylation factor-like protein 5B | Mus musculus (Mouse) | SS |
| P84078 | Arf1 | ADP-ribosylation factor 1 | Mus musculus (Mouse) | SS |
| Q8BSL7 | Arf2 | ADP-ribosylation factor 2 | Mus musculus (Mouse) | SS |
| P61750 | Arf4 | ADP-ribosylation factor 4 | Mus musculus (Mouse) | SS |
| P51824 | ADP-ribosylation factor 1 | Solanum tuberosum (Potato) | SS | |
| P84082 | Arf2 | ADP-ribosylation factor 2 | Rattus norvegicus (Rat) | SS |
| P51646 | Arl5a | ADP-ribosylation factor-like protein 5A | Rattus norvegicus (Rat) | SS |
| P84083 | Arf5 | ADP-ribosylation factor 5 | Rattus norvegicus (Rat) | SS |
| P36407 | Trim23 | E3 ubiquitin-protein ligase TRIM23 | Rattus norvegicus (Rat) | SS |
| P61206 | Arf3 | ADP-ribosylation factor 3 | Rattus norvegicus (Rat) | SS |
| P84079 | Arf1 | ADP-ribosylation factor 1 | Rattus norvegicus (Rat) | SS |
| P61751 | Arf4 | ADP-ribosylation factor 4 | Rattus norvegicus (Rat) | SS |
| P51823 | ARF | ADP-ribosylation factor 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q06396 | Os01g0813400 | ADP-ribosylation factor 1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10943 | arf-1.2 | ADP-ribosylation factor 1-like 2 | Caenorhabditis elegans | SS |
| P34212 | arl-5 | ADP-ribosylation factor-like protein 5 | Caenorhabditis elegans | SS |
| P40940 | ARF3 | ADP-ribosylation factor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P36397 | ARF1 | ADP-ribosylation factor 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P0DH91 | ARF2-B | ADP-ribosylation factor 2-B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LQC8 | ARF2-A | ADP-ribosylation factor 2-A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5M9P8 | arl6 | ADP-ribosylation factor-like protein 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGLTISSLFS | RLFGKKQMRI | LMVGLDAAGK | TTILYKLKLG | EIVTTIPTIG | FNVETVEYKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ICFTVWDVGG | QDRIRPLWKH | YFQNTQGLIF | VVDSNDRERI | QEVADELQKM | LLVDELRDAV |
| 130 | 140 | 150 | 160 | 170 | |
| LLLFANKQDL | PNAMAISEMT | DKLGLQSLRN | RTWYVQATCA | TQGTGLYEGL | DWLSNELSKR |