P18031
Gene name |
PTPN1 (PTP1B) |
Protein name |
Tyrosine-protein phosphatase non-receptor type 1 |
Names |
Protein-tyrosine phosphatase 1B, PTP-1B |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5770 |
EC number |
3.1.3.48: Phosphoric monoester hydrolases |
Protein Class |
|
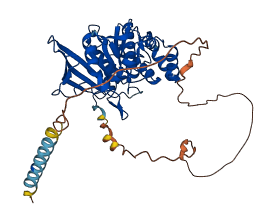
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

429 structures for P18031
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1A5Y | X-ray | 250 A | A | 1-330 | PDB |
| 1AAX | X-ray | 190 A | A | 1-321 | PDB |
| 1BZC | X-ray | 235 A | A | 1-321 | PDB |
| 1BZH | X-ray | 210 A | A | 1-298 | PDB |
| 1BZJ | X-ray | 225 A | A | 2-298 | PDB |
| 1C83 | X-ray | 180 A | A | 1-298 | PDB |
| 1C84 | X-ray | 235 A | A | 1-298 | PDB |
| 1C85 | X-ray | 272 A | A | 1-298 | PDB |
| 1C86 | X-ray | 230 A | A | 1-298 | PDB |
| 1C87 | X-ray | 210 A | A | 1-298 | PDB |
| 1C88 | X-ray | 180 A | A | 1-298 | PDB |
| 1ECV | X-ray | 195 A | A | 1-298 | PDB |
| 1EEN | X-ray | 190 A | A | 1-321 | PDB |
| 1EEO | X-ray | 180 A | A | 1-321 | PDB |
| 1G1F | X-ray | 200 A | A | 1-298 | PDB |
| 1G1G | X-ray | 220 A | A | 1-298 | PDB |
| 1G1H | X-ray | 240 A | A | 1-298 | PDB |
| 1G7F | X-ray | 180 A | A | 1-298 | PDB |
| 1G7G | X-ray | 220 A | A | 1-298 | PDB |
| 1GFY | X-ray | 213 A | A | 1-298 | PDB |
| 1I57 | X-ray | 210 A | A | 1-298 | PDB |
| 1JF7 | X-ray | 220 A | A/B | 1-298 | PDB |
| 1KAK | X-ray | 250 A | A | 1-298 | PDB |
| 1KAV | X-ray | 235 A | A | 1-298 | PDB |
| 1L8G | X-ray | 250 A | A | 1-321 | PDB |
| 1LQF | X-ray | 250 A | A/B/C/D | 1-283 | PDB |
| 1NL9 | X-ray | 240 A | A | 1-321 | PDB |
| 1NNY | X-ray | 240 A | A | 1-321 | PDB |
| 1NO6 | X-ray | 240 A | A | 1-321 | PDB |
| 1NWE | X-ray | 310 A | A | 1-298 | PDB |
| 1NWL | X-ray | 240 A | A | 1-298 | PDB |
| 1NZ7 | X-ray | 240 A | A | 1-321 | PDB |
| 1OEM | X-ray | 180 A | X | 1-321 | PDB |
| 1OEO | X-ray | 215 A | X | 1-321 | PDB |
| 1OES | X-ray | 220 A | A | 1-321 | PDB |
| 1OET | X-ray | 230 A | A | 1-321 | PDB |
| 1OEU | X-ray | 250 A | A | 1-321 | PDB |
| 1OEV | X-ray | 220 A | A | 1-321 | PDB |
| 1ONY | X-ray | 215 A | A | 1-321 | PDB |
| 1ONZ | X-ray | 240 A | A | 1-321 | PDB |
| 1PA1 | X-ray | 160 A | A | 1-298 | PDB |
| 1PH0 | X-ray | 220 A | A | 1-321 | PDB |
| 1PTT | X-ray | 290 A | A | 1-321 | PDB |
| 1PTU | X-ray | 260 A | A | 1-321 | PDB |
| 1PTV | X-ray | 230 A | A | 1-321 | PDB |
| 1PTY | X-ray | 185 A | A | 1-321 | PDB |
| 1PXH | X-ray | 215 A | A | 1-321 | PDB |
| 1PYN | X-ray | 220 A | A | 1-321 | PDB |
| 1Q1M | X-ray | 260 A | A | 1-321 | PDB |
| 1Q6J | X-ray | 220 A | A | 1-298 | PDB |
| 1Q6M | X-ray | 220 A | A | 1-298 | PDB |
| 1Q6N | X-ray | 210 A | A/B | 1-298 | PDB |
| 1Q6P | X-ray | 230 A | A/B | 1-298 | PDB |
| 1Q6S | X-ray | 220 A | A/B | 1-298 | PDB |
| 1Q6T | X-ray | 230 A | A/B | 1-298 | PDB |
| 1QXK | X-ray | 230 A | A | 1-321 | PDB |
| 1SUG | X-ray | 195 A | A | 1-321 | PDB |
| 1T48 | X-ray | 220 A | A | 1-298 | PDB |
| 1T49 | X-ray | 190 A | A | 1-298 | PDB |
| 1T4J | X-ray | 270 A | A | 1-298 | PDB |
| 1WAX | X-ray | 220 A | A | 1-321 | PDB |
| 1XBO | X-ray | 250 A | A | 1-321 | PDB |
| 2AZR | X-ray | 200 A | A | 1-299 | PDB |
| 2B07 | X-ray | 210 A | A | 1-299 | PDB |
| 2B4S | X-ray | 230 A | A/C | 1-298 | PDB |
| 2BGD | X-ray | 240 A | A | 1-321 | PDB |
| 2BGE | X-ray | 180 A | A | 1-321 | PDB |
| 2CM2 | X-ray | 150 A | A | 2-298 | PDB |
| 2CM3 | X-ray | 210 A | A/B | 1-298 | PDB |
| 2CM7 | X-ray | 210 A | A | 1-321 | PDB |
| 2CM8 | X-ray | 210 A | A | 1-321 | PDB |
| 2CMA | X-ray | 230 A | A | 1-321 | PDB |
| 2CMB | X-ray | 170 A | A | 1-298 | PDB |
| 2CMC | X-ray | 220 A | A | 1-298 | PDB |
| 2CNE | X-ray | 180 A | A | 1-298 | PDB |
| 2CNF | X-ray | 220 A | A | 1-321 | PDB |
| 2CNG | X-ray | 190 A | A | 1-321 | PDB |
| 2CNH | X-ray | 180 A | A | 1-321 | PDB |
| 2CNI | X-ray | 200 A | A | 1-321 | PDB |
| 2F6F | X-ray | 200 A | A | 1-298 | PDB |
| 2F6T | X-ray | 170 A | A | 1-298 | PDB |
| 2F6V | X-ray | 170 A | A | 1-298 | PDB |
| 2F6W | X-ray | 220 A | A | 1-298 | PDB |
| 2F6Y | X-ray | 215 A | A | 1-298 | PDB |
| 2F6Z | X-ray | 170 A | A | 1-298 | PDB |
| 2F70 | X-ray | 212 A | A | 1-298 | PDB |
| 2F71 | X-ray | 155 A | A | 1-298 | PDB |
| 2FJM | X-ray | 210 A | A/B | 1-298 | PDB |
| 2FJN | X-ray | 220 A | A/B | 1-298 | PDB |
| 2H4G | X-ray | 250 A | A | 1-299 | PDB |
| 2H4K | X-ray | 230 A | A | 1-299 | PDB |
| 2HB1 | X-ray | 200 A | A | 1-299 | PDB |
| 2HNP | X-ray | 285 A | A | 1-321 | PDB |
| 2HNQ | X-ray | 285 A | A | 1-321 | PDB |
| 2NT7 | X-ray | 210 A | A | 1-299 | PDB |
| 2NTA | X-ray | 210 A | A | 1-299 | PDB |
| 2QBP | X-ray | 250 A | A | 1-299 | PDB |
| 2QBQ | X-ray | 210 A | A | 1-299 | PDB |
| 2QBR | X-ray | 230 A | A | 1-299 | PDB |
| 2QBS | X-ray | 210 A | A | 1-299 | PDB |
| 2VEU | X-ray | 240 A | A | 1-321 | PDB |
| 2VEV | X-ray | 180 A | A | 1-321 | PDB |
| 2VEW | X-ray | 200 A | A | 1-321 | PDB |
| 2VEX | X-ray | 220 A | A | 1-321 | PDB |
| 2VEY | X-ray | 220 A | A | 1-321 | PDB |
| 2ZMM | X-ray | 210 A | A | 1-299 | PDB |
| 2ZN7 | X-ray | 210 A | A | 1-299 | PDB |
| 3A5J | X-ray | 170 A | A | 2-321 | PDB |
| 3A5K | X-ray | 185 A | A | 2-298 | PDB |
| 3CWE | X-ray | 160 A | A | 1-283 | PDB |
| 3D9C | X-ray | 230 A | A | 1-321 | PDB |
| 3EAX | X-ray | 190 A | A | 1-321 | PDB |
| 3EB1 | X-ray | 240 A | A | 1-321 | PDB |
| 3EU0 | X-ray | 270 A | A | 1-282 | PDB |
| 3I7Z | X-ray | 230 A | A | 1-321 | PDB |
| 3I80 | X-ray | 225 A | A | 1-321 | PDB |
| 3QKP | X-ray | 205 A | A | 1-321 | PDB |
| 3QKQ | X-ray | 220 A | A | 1-321 | PDB |
| 3SME | X-ray | 170 A | A | 1-298 | PDB |
| 3ZMP | X-ray | 262 A | A/B | 1-321 | PDB |
| 3ZMQ | X-ray | 330 A | A | 1-321 | PDB |
| 3ZV2 | X-ray | 280 A | A | 1-320 | PDB |
| 4BJO | X-ray | 206 A | A/B | 2-321 | PDB |
| 4I8N | X-ray | 250 A | A | 1-320 | PDB |
| 4QAH | X-ray | 240 A | A | 1-299 | PDB |
| 4QAP | X-ray | 190 A | A | 1-299 | PDB |
| 4QBE | X-ray | 229 A | A | 1-298 | PDB |
| 4QBW | X-ray | 191 A | A | 1-299 | PDB |
| 4Y14 | X-ray | 190 A | A/B | 2-301 | PDB |
| 4ZRT | X-ray | 174 A | A | 1-298 | PDB |
| 5K9V | X-ray | 190 A | A | 1-301 | PDB |
| 5K9W | X-ray | 201 A | A | 1-301 | PDB |
| 5KA0 | X-ray | 199 A | A | 1-284 | PDB |
| 5KA1 | X-ray | 184 A | A | 1-284 | PDB |
| 5KA2 | X-ray | 207 A | A | 1-301 | PDB |
| 5KA3 | X-ray | 214 A | A | 1-301 | PDB |
| 5KA4 | X-ray | 219 A | A | 1-301 | PDB |
| 5KA7 | X-ray | 206 A | A | 1-301 | PDB |
| 5KA8 | X-ray | 197 A | A | 1-301 | PDB |
| 5KA9 | X-ray | 207 A | A | 1-301 | PDB |
| 5KAA | X-ray | 197 A | A | 1-284 | PDB |
| 5KAB | X-ray | 197 A | A | 1-284 | PDB |
| 5KAC | X-ray | 190 A | A | 1-301 | PDB |
| 5KAD | X-ray | 190 A | A/B | 1-301 | PDB |
| 5QDE | X-ray | 176 A | A | 1-321 | PDB |
| 5QDF | X-ray | 171 A | A | 1-321 | PDB |
| 5QDG | X-ray | 179 A | A | 1-321 | PDB |
| 5QDH | X-ray | 168 A | A | 1-321 | PDB |
| 5QDI | X-ray | 162 A | A | 1-321 | PDB |
| 5QDJ | X-ray | 176 A | A | 1-321 | PDB |
| 5QDK | X-ray | 155 A | A | 1-321 | PDB |
| 5QDL | X-ray | 183 A | A | 1-321 | PDB |
| 5QDM | X-ray | 265 A | A | 1-321 | PDB |
| 5QDN | X-ray | 182 A | A | 1-321 | PDB |
| 5QDO | X-ray | 179 A | A | 1-321 | PDB |
| 5QDP | X-ray | 174 A | A | 1-321 | PDB |
| 5QDQ | X-ray | 157 A | A | 1-321 | PDB |
| 5QDR | X-ray | 178 A | A | 1-321 | PDB |
| 5QDS | X-ray | 175 A | A | 1-321 | PDB |
| 5QDT | X-ray | 182 A | A | 1-321 | PDB |
| 5QDU | X-ray | 167 A | A | 1-321 | PDB |
| 5QDV | X-ray | 177 A | A | 1-321 | PDB |
| 5QDW | X-ray | 221 A | A | 1-321 | PDB |
| 5QDX | X-ray | 206 A | A | 1-321 | PDB |
| 5QDY | X-ray | 183 A | A | 1-321 | PDB |
| 5QDZ | X-ray | 214 A | A | 1-321 | PDB |
| 5QE0 | X-ray | 198 A | A | 1-321 | PDB |
| 5QE1 | X-ray | 169 A | A | 1-321 | PDB |
| 5QE2 | X-ray | 179 A | A | 1-321 | PDB |
| 5QE3 | X-ray | 174 A | A | 1-321 | PDB |
| 5QE4 | X-ray | 185 A | A | 1-321 | PDB |
| 5QE5 | X-ray | 177 A | A | 1-321 | PDB |
| 5QE6 | X-ray | 177 A | A | 1-321 | PDB |
| 5QE7 | X-ray | 171 A | A | 1-321 | PDB |
| 5QE8 | X-ray | 181 A | A | 1-321 | PDB |
| 5QE9 | X-ray | 169 A | A | 1-321 | PDB |
| 5QEA | X-ray | 174 A | A | 1-321 | PDB |
| 5QEB | X-ray | 173 A | A | 1-321 | PDB |
| 5QEC | X-ray | 167 A | A | 1-321 | PDB |
| 5QED | X-ray | 175 A | A | 1-321 | PDB |
| 5QEE | X-ray | 193 A | A | 1-321 | PDB |
| 5QEF | X-ray | 160 A | A | 1-321 | PDB |
| 5QEG | X-ray | 197 A | A | 1-321 | PDB |
| 5QEH | X-ray | 194 A | A | 1-321 | PDB |
| 5QEI | X-ray | 174 A | A | 1-321 | PDB |
| 5QEJ | X-ray | 192 A | A | 1-321 | PDB |
| 5QEK | X-ray | 190 A | A | 1-321 | PDB |
| 5QEL | X-ray | 165 A | A | 1-321 | PDB |
| 5QEM | X-ray | 175 A | A | 1-321 | PDB |
| 5QEN | X-ray | 177 A | A | 1-321 | PDB |
| 5QEO | X-ray | 172 A | A | 1-321 | PDB |
| 5QEP | X-ray | 177 A | A | 1-321 | PDB |
| 5QEQ | X-ray | 197 A | A | 1-321 | PDB |
| 5QER | X-ray | 173 A | A | 1-321 | PDB |
| 5QES | X-ray | 175 A | A | 1-321 | PDB |
| 5QET | X-ray | 172 A | A | 1-321 | PDB |
| 5QEU | X-ray | 174 A | A | 1-321 | PDB |
| 5QEV | X-ray | 172 A | A | 1-321 | PDB |
| 5QEW | X-ray | 183 A | A | 1-321 | PDB |
| 5QEX | X-ray | 167 A | A | 1-321 | PDB |
| 5QEY | X-ray | 177 A | A | 1-321 | PDB |
| 5QEZ | X-ray | 165 A | A | 1-321 | PDB |
| 5QF0 | X-ray | 171 A | A | 1-321 | PDB |
| 5QF1 | X-ray | 184 A | A | 1-321 | PDB |
| 5QF2 | X-ray | 177 A | A | 1-321 | PDB |
| 5QF3 | X-ray | 156 A | A | 1-321 | PDB |
| 5QF4 | X-ray | 183 A | A | 1-321 | PDB |
| 5QF5 | X-ray | 173 A | A | 1-321 | PDB |
| 5QF6 | X-ray | 176 A | A | 1-321 | PDB |
| 5QF7 | X-ray | 175 A | A | 1-321 | PDB |
| 5QF8 | X-ray | 199 A | A | 1-321 | PDB |
| 5QF9 | X-ray | 194 A | A | 1-321 | PDB |
| 5QFA | X-ray | 174 A | A | 1-321 | PDB |
| 5QFB | X-ray | 185 A | A | 1-321 | PDB |
| 5QFC | X-ray | 184 A | A | 1-321 | PDB |
| 5QFD | X-ray | 169 A | A | 1-321 | PDB |
| 5QFE | X-ray | 156 A | A | 1-321 | PDB |
| 5QFF | X-ray | 170 A | A | 1-321 | PDB |
| 5QFG | X-ray | 165 A | A | 1-321 | PDB |
| 5QFH | X-ray | 159 A | A | 1-321 | PDB |
| 5QFI | X-ray | 168 A | A | 1-321 | PDB |
| 5QFJ | X-ray | 195 A | A | 1-321 | PDB |
| 5QFK | X-ray | 159 A | A | 1-321 | PDB |
| 5QFL | X-ray | 182 A | A | 1-321 | PDB |
| 5QFM | X-ray | 183 A | A | 1-321 | PDB |
| 5QFN | X-ray | 168 A | A | 1-321 | PDB |
| 5QFO | X-ray | 185 A | A | 1-321 | PDB |
| 5QFP | X-ray | 177 A | A | 1-321 | PDB |
| 5QFQ | X-ray | 162 A | A | 1-321 | PDB |
| 5QFR | X-ray | 162 A | A | 1-321 | PDB |
| 5QFS | X-ray | 185 A | A | 1-321 | PDB |
| 5QFT | X-ray | 196 A | A | 1-321 | PDB |
| 5QFU | X-ray | 161 A | A | 1-321 | PDB |
| 5QFV | X-ray | 164 A | A | 1-321 | PDB |
| 5QFW | X-ray | 166 A | A | 1-321 | PDB |
| 5QFX | X-ray | 182 A | A | 1-321 | PDB |
| 5QFY | X-ray | 177 A | A | 1-321 | PDB |
| 5QFZ | X-ray | 172 A | A | 1-321 | PDB |
| 5QG0 | X-ray | 175 A | A | 1-321 | PDB |
| 5QG1 | X-ray | 221 A | A | 1-321 | PDB |
| 5QG2 | X-ray | 212 A | A | 1-321 | PDB |
| 5QG3 | X-ray | 165 A | A | 1-321 | PDB |
| 5QG4 | X-ray | 179 A | A | 1-321 | PDB |
| 5QG5 | X-ray | 207 A | A | 1-321 | PDB |
| 5QG6 | X-ray | 173 A | A | 1-321 | PDB |
| 5QG7 | X-ray | 181 A | A | 1-321 | PDB |
| 5QG8 | X-ray | 163 A | A | 1-321 | PDB |
| 5QG9 | X-ray | 167 A | A | 1-321 | PDB |
| 5QGA | X-ray | 165 A | A | 1-321 | PDB |
| 5QGB | X-ray | 155 A | A | 1-321 | PDB |
| 5QGC | X-ray | 159 A | A | 1-321 | PDB |
| 5QGD | X-ray | 166 A | A | 1-321 | PDB |
| 5QGE | X-ray | 170 A | A | 1-321 | PDB |
| 5QGF | X-ray | 151 A | A | 1-321 | PDB |
| 5T19 | X-ray | 210 A | A | 1-321 | PDB |
| 6B8E | X-ray | 182 A | A | 1-321 | PDB |
| 6B8T | X-ray | 185 A | A | 1-321 | PDB |
| 6B8X | X-ray | 174 A | A | 1-321 | PDB |
| 6B8Z | X-ray | 180 A | A | 1-321 | PDB |
| 6B90 | X-ray | 195 A | A | 1-321 | PDB |
| 6B95 | X-ray | 195 A | A | 1-321 | PDB |
| 6BAI | X-ray | 195 A | A | 1-321 | PDB |
| 6CWU | X-ray | 208 A | A | 1-321 | PDB |
| 6CWV | X-ray | 198 A | A | 1-321 | PDB |
| 6NTP | X-ray | 189 A | A | 1-298 | PDB |
| 6OL4 | X-ray | 215 A | A | 2-297 | PDB |
| 6OLQ | X-ray | 210 A | A | 2-298 | PDB |
| 6OLV | X-ray | 210 A | A | 2-297 | PDB |
| 6OMY | X-ray | 210 A | A | 2-298 | PDB |
| 6PFW | X-ray | 234 A | A | 2-298 | PDB |
| 6PG0 | X-ray | 210 A | A | 2-298 | PDB |
| 6PGT | X-ray | 220 A | A | 2-299 | PDB |
| 6PHA | X-ray | 230 A | A | 2-298 | PDB |
| 6PHS | X-ray | 213 A | A | 2-298 | PDB |
| 6PM8 | X-ray | 206 A | A | 2-298 | PDB |
| 6W30 | X-ray | 210 A | A | 1-321 | PDB |
| 6XE8 | X-ray | 195 A | A | 1-321 | PDB |
| 6XEA | X-ray | 155 A | A | 1-321 | PDB |
| 6XED | X-ray | 179 A | A | 1-321 | PDB |
| 6XEE | X-ray | 250 A | A | 1-321 | PDB |
| 6XEF | X-ray | 205 A | A | 1-321 | PDB |
| 6XEG | X-ray | 255 A | A | 1-321 | PDB |
| 7FQM | X-ray | 194 A | A | 1-321 | PDB |
| 7FQN | X-ray | 204 A | A | 1-321 | PDB |
| 7FQO | X-ray | 193 A | A | 1-321 | PDB |
| 7FQP | X-ray | 188 A | A | 1-321 | PDB |
| 7FQQ | X-ray | 188 A | A | 1-321 | PDB |
| 7FQR | X-ray | 190 A | A | 1-321 | PDB |
| 7FQS | X-ray | 212 A | A | 1-321 | PDB |
| 7FQT | X-ray | 254 A | A | 1-321 | PDB |
| 7FQU | X-ray | 186 A | A | 1-321 | PDB |
| 7FQV | X-ray | 204 A | A | 1-321 | PDB |
| 7FQW | X-ray | 217 A | A | 1-321 | PDB |
| 7FQX | X-ray | 246 A | A | 1-321 | PDB |
| 7FQY | X-ray | 213 A | A | 1-321 | PDB |
| 7FQZ | X-ray | 209 A | A | 1-321 | PDB |
| 7FRE | X-ray | 185 A | A | 1-321 | PDB |
| 7FRF | X-ray | 215 A | A | 1-321 | PDB |
| 7FRG | X-ray | 184 A | A | 1-321 | PDB |
| 7FRH | X-ray | 184 A | A | 1-321 | PDB |
| 7FRI | X-ray | 186 A | A | 1-321 | PDB |
| 7FRJ | X-ray | 180 A | A | 1-321 | PDB |
| 7FRK | X-ray | 180 A | A | 1-321 | PDB |
| 7FRL | X-ray | 179 A | A | 1-321 | PDB |
| 7FRM | X-ray | 191 A | A | 1-321 | PDB |
| 7FRN | X-ray | 185 A | A | 1-321 | PDB |
| 7FRO | X-ray | 193 A | A | 1-321 | PDB |
| 7FRP | X-ray | 177 A | A | 1-321 | PDB |
| 7FRQ | X-ray | 201 A | A | 1-321 | PDB |
| 7FRR | X-ray | 183 A | A | 1-321 | PDB |
| 7FRS | X-ray | 183 A | A | 1-321 | PDB |
| 7FRT | X-ray | 186 A | A | 1-321 | PDB |
| 7FRU | X-ray | 198 A | A | 1-321 | PDB |
| 7GS7 | X-ray | 166 A | A | 1-321 | PDB |
| 7GS8 | X-ray | 167 A | A | 1-321 | PDB |
| 7GS9 | X-ray | 196 A | A | 1-321 | PDB |
| 7GSA | X-ray | 172 A | A | 1-321 | PDB |
| 7GSB | X-ray | 172 A | A | 1-321 | PDB |
| 7GSC | X-ray | 169 A | A | 1-321 | PDB |
| 7GSD | X-ray | 180 A | A | 1-321 | PDB |
| 7GSE | X-ray | 189 A | A | 1-321 | PDB |
| 7GSF | X-ray | 178 A | A | 1-321 | PDB |
| 7GSG | X-ray | 190 A | A | 1-321 | PDB |
| 7GSH | X-ray | 188 A | A | 1-321 | PDB |
| 7GSI | X-ray | 171 A | A | 1-321 | PDB |
| 7GSJ | X-ray | 177 A | A | 1-321 | PDB |
| 7GSK | X-ray | 184 A | A | 1-321 | PDB |
| 7GSL | X-ray | 177 A | A | 1-321 | PDB |
| 7GSM | X-ray | 203 A | A | 1-321 | PDB |
| 7GSN | X-ray | 187 A | A | 1-321 | PDB |
| 7GSO | X-ray | 172 A | A | 1-321 | PDB |
| 7GSQ | X-ray | 173 A | A | 1-321 | PDB |
| 7GSR | X-ray | 169 A | A | 1-321 | PDB |
| 7GST | X-ray | 164 A | A | 1-321 | PDB |
| 7GSU | X-ray | 165 A | A | 1-321 | PDB |
| 7GSV | X-ray | 192 A | A | 1-321 | PDB |
| 7GSW | X-ray | 179 A | A | 1-321 | PDB |
| 7GSX | X-ray | 167 A | A | 1-321 | PDB |
| 7GSY | X-ray | 197 A | A | 1-321 | PDB |
| 7GSZ | X-ray | 191 A | A | 1-321 | PDB |
| 7GT0 | X-ray | 176 A | A | 1-321 | PDB |
| 7GT1 | X-ray | 191 A | A | 1-321 | PDB |
| 7GT2 | X-ray | 190 A | A | 1-321 | PDB |
| 7GT3 | X-ray | 159 A | A | 1-321 | PDB |
| 7GT4 | X-ray | 175 A | A | 1-321 | PDB |
| 7GT5 | X-ray | 161 A | A | 1-321 | PDB |
| 7GT6 | X-ray | 166 A | A | 1-321 | PDB |
| 7GT7 | X-ray | 184 A | A | 1-321 | PDB |
| 7GT8 | X-ray | 191 A | A | 1-321 | PDB |
| 7GT9 | X-ray | 190 A | A | 1-321 | PDB |
| 7GTA | X-ray | 206 A | A | 1-321 | PDB |
| 7GTB | X-ray | 186 A | A | 1-321 | PDB |
| 7GTC | X-ray | 192 A | A | 1-321 | PDB |
| 7GTD | X-ray | 191 A | A | 1-321 | PDB |
| 7GTE | X-ray | 190 A | A | 1-321 | PDB |
| 7GTF | X-ray | 197 A | A | 1-321 | PDB |
| 7GTG | X-ray | 169 A | A | 1-321 | PDB |
| 7GTH | X-ray | 166 A | A | 1-321 | PDB |
| 7GTI | X-ray | 166 A | A | 1-321 | PDB |
| 7GTJ | X-ray | 183 A | A | 1-321 | PDB |
| 7GTK | X-ray | 176 A | A | 1-321 | PDB |
| 7GTL | X-ray | 183 A | A | 1-321 | PDB |
| 7GTM | X-ray | 172 A | A | 1-321 | PDB |
| 7GTN | X-ray | 166 A | A | 1-321 | PDB |
| 7GTO | X-ray | 165 A | A | 1-321 | PDB |
| 7GTP | X-ray | 247 A | A | 1-321 | PDB |
| 7GTQ | X-ray | 209 A | A | 1-321 | PDB |
| 7GTR | X-ray | 178 A | A | 1-321 | PDB |
| 7GTS | X-ray | 180 A | A | 1-321 | PDB |
| 7GTT | X-ray | 193 A | A | 1-321 | PDB |
| 7GTU | X-ray | 208 A | A | 1-321 | PDB |
| 7GTV | X-ray | 172 A | A | 1-321 | PDB |
| 7GTW | X-ray | 151 A | A | 1-321 | PDB |
| 7GTX | X-ray | 151 A | A | 1-321 | PDB |
| 7GTY | X-ray | 154 A | A | 1-321 | PDB |
| 7GTZ | X-ray | 160 A | A | 1-321 | PDB |
| 7GU0 | X-ray | 166 A | A | 1-321 | PDB |
| 7GU1 | X-ray | 159 A | A | 1-321 | PDB |
| 7GU2 | X-ray | 180 A | A | 1-321 | PDB |
| 7GU3 | X-ray | 174 A | A | 1-321 | PDB |
| 7GU4 | X-ray | 159 A | A | 1-321 | PDB |
| 7GU5 | X-ray | 152 A | A | 1-321 | PDB |
| 7GU6 | X-ray | 156 A | A | 1-321 | PDB |
| 7GU7 | X-ray | 170 A | A | 1-321 | PDB |
| 7GU8 | X-ray | 159 A | A | 1-321 | PDB |
| 7GU9 | X-ray | 153 A | A | 1-321 | PDB |
| 7GUA | X-ray | 163 A | A | 1-321 | PDB |
| 7GUB | X-ray | 194 A | A | 1-321 | PDB |
| 7GUC | X-ray | 178 A | A | 1-321 | PDB |
| 7KEN | X-ray | 180 A | A | 2-323 | PDB |
| 7KEY | X-ray | 177 A | A | 2-283 | PDB |
| 7KLX | X-ray | 184 A | A | 2-283 | PDB |
| 7L0C | X-ray | 180 A | A | 1-321 | PDB |
| 7L0H | X-ray | 210 A | A | 1-321 | PDB |
| 7LFO | X-ray | 194 A | A | 1-321 | PDB |
| 7MKZ | X-ray | 140 A | A | 1-301 | PDB |
| 7MM1 | X-ray | 185 A | A | 1-321 | PDB |
| 7MN7 | X-ray | 195 A | A | 1-301 | PDB |
| 7MN9 | X-ray | 124 A | A | 1-284 | PDB |
| 7MNA | X-ray | 147 A | A | 1-284 | PDB |
| 7MNB | X-ray | 220 A | A | 1-301 | PDB |
| 7MNC | X-ray | 185 A | A | 1-301 | PDB |
| 7MND | X-ray | 229 A | A | 1-301 | PDB |
| 7MNE | X-ray | 160 A | A | 1-301 | PDB |
| 7MNF | X-ray | 170 A | A | 1-301 | PDB |
| 7MOU | X-ray | 148 A | A | 1-284 | PDB |
| 7MOV | X-ray | 165 A | A/B | 1-301 | PDB |
| 7MOW | X-ray | 180 A | A | 1-301 | PDB |
| 7RIN | X-ray | 185 A | A | 1-321 | PDB |
| 7S4F | X-ray | 165 A | A | 1-298 | PDB |
| 8DU7 | X-ray | 240 A | A | 1-284 | PDB |
| 8EXI | X-ray | 160 A | A | 1-297 | PDB |
| 8EXJ | X-ray | 230 A | A | 1-299 | PDB |
| 8EXK | X-ray | 210 A | A | 3-299 | PDB |
| 8EXM | X-ray | 235 A | A | 1-299 | PDB |
| 8EXN | X-ray | 215 A | A | 1-299 | PDB |
| 8EYA | X-ray | 210 A | A/B | 1-301 | PDB |
| 8EYB | X-ray | 235 A | A/B | 2-297 | PDB |
| 8EYC | X-ray | 299 A | A | 1-299 | PDB |
| 8F88 | X-ray | 310 A | A/B/C | 1-321 | PDB |
| 8G65 | X-ray | 145 A | A/B | 1-298 | PDB |
| 8G67 | X-ray | 153 A | A/B | 1-298 | PDB |
| 8G68 | X-ray | 182 A | A/B | 1-298 | PDB |
| 8G69 | X-ray | 153 A | A/B | 1-298 | PDB |
| 8G6A | X-ray | 162 A | A/B | 1-298 | PDB |
| 8SKL | X-ray | 155 A | A | 1-321 | PDB |
| 8U1E | X-ray | 143 A | A/B | 1-321 | PDB |
| 8XOY | X-ray | 155 A | A | 1-321 | PDB |
| AF-P18031-F1 | Predicted | AlphaFoldDB |
237 variants for P18031
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1407157199 CA408968464 |
2 | E>V | No |
ClinGen TOPMed |
|
|
rs755655338 CA315210982 |
5 | K>N | No |
ClinGen Ensembl |
|
|
CA315210983 rs148137197 |
6 | E>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1489441940 CA408968635 |
7 | F>L | No |
ClinGen gnomAD |
|
|
CA408968695 rs1258528842 |
9 | Q>R | No |
ClinGen TOPMed |
|
|
CA408968731 rs1246414151 |
10 | I>N | No |
ClinGen gnomAD |
|
|
CA408968745 rs1487620720 |
11 | D>Y | No |
ClinGen gnomAD |
|
|
CA315210984 rs925151133 |
12 | K>R | No |
ClinGen Ensembl |
|
|
CA9906383 rs748228918 |
13 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1401777841 CA408968885 |
15 | S>N | No |
ClinGen Ensembl |
|
|
CA9906385 rs777863121 |
15 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA408968957 rs1381957080 |
18 | A>V | No |
ClinGen gnomAD |
|
|
CA9906386 rs150635098 |
19 | I>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA408962618 rs1443524226 |
24 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs545064112 CA9906397 |
25 | H>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs751952672 CA9906398 |
26 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA315194068 rs201758338 |
28 | S>N | No |
ClinGen 1000Genomes TOPMed |
|
|
rs201758338 CA408962645 |
28 | S>T | No |
ClinGen 1000Genomes TOPMed |
|
|
rs755602646 CA9906399 |
29 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs759320660 CA315194085 |
43 | R>* | No |
ClinGen Ensembl |
|
| TCGA novel | 43 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 44 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA315194090 rs767518093 |
46 | Y>* | No |
ClinGen Ensembl |
|
|
CA9906401 rs753192131 |
47 | R>K | No |
ClinGen ExAC gnomAD |
|
|
CA9906403 rs777861433 |
49 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA408962796 rs1473624032 |
50 | S>C | No |
ClinGen gnomAD |
|
|
rs1267779326 CA408962797 |
50 | S>N | No |
ClinGen TOPMed |
|
| TCGA novel | 51 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs753938375 CA9906424 |
54 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA315196010 rs753938375 |
54 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs779200250 CA9906426 |
57 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA315196035 rs746039629 |
63 | D>V | No |
ClinGen Ensembl |
|
| TCGA novel | 69 | A>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 69 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 70 | S>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9906428 rs754785542 |
76 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs992322197 CA315196047 |
76 | E>K | No |
ClinGen TOPMed |
|
|
rs780939045 CA9906429 |
78 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408963029 rs1273120831 |
80 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA408963062 rs1285079073 |
85 | Q>R | No |
ClinGen gnomAD |
|
| TCGA novel | 88 | L>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1217160644 CA408963103 |
89 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs189361363 CA9906450 |
92 | C>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs770497491 CA9906452 |
93 | G>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA9906453 rs770497491 |
93 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1330974189 CA408963175 |
99 | V>A | No |
ClinGen TOPMed |
|
|
CA408963215 rs1177454007 |
104 | S>I | No |
ClinGen gnomAD |
|
|
rs775223257 CA9906456 |
108 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA408963265 rs1364690180 |
112 | R>G | No |
ClinGen gnomAD |
|
|
CA9906457 rs375554144 |
114 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs768861855 CA9906458 |
115 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9906459 rs777216811 |
116 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA408963312 rs1173031559 |
118 | S>L | No |
ClinGen TOPMed |
|
|
CA9906473 rs779882276 |
122 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA408963393 rs1430775714 |
128 | K>E | No |
ClinGen TOPMed |
|
|
CA408963436 rs1414699318 |
133 | M>T | No |
ClinGen gnomAD |
|
| TCGA novel | 134 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA408963486 rs1601413769 |
140 | L>M | No |
ClinGen Ensembl |
|
|
CA408963494 rs1209225422 |
141 | K>E | No |
ClinGen gnomAD |
|
|
CA9906478 rs762446257 |
149 | I>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762446257 CA9906477 |
149 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408963551 rs1475678011 |
149 | I>V | No |
ClinGen gnomAD |
|
|
rs1314516315 CA408963634 |
161 | E>G | No |
ClinGen gnomAD |
|
|
rs1392325234 CA408963654 |
164 | T>A | No |
ClinGen Ensembl |
|
|
rs1272267070 CA408963656 |
164 | T>I | No |
ClinGen TOPMed |
|
|
CA9906505 rs149080433 |
165 | T>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA408964167 rs1349231783 |
166 | Q>K | No |
ClinGen TOPMed gnomAD |
|
|
rs529107431 CA315204335 |
166 | Q>P | No |
ClinGen Ensembl |
|
|
rs762225731 CA408964175 |
167 | E>A | No |
ClinGen gnomAD |
|
|
CA408964177 rs1568797402 |
167 | E>D | No |
ClinGen Ensembl |
|
|
rs762225731 CA315204342 |
167 | E>G | No |
ClinGen gnomAD |
|
|
rs761728507 CA9906506 |
168 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs765072009 CA9906507 |
169 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9906508 rs750431969 |
171 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1195564673 CA408964259 |
179 | W>* | No |
ClinGen gnomAD |
|
|
CA9906511 rs372483862 |
180 | P>S | No |
ClinGen ESP ExAC gnomAD |
|
| TCGA novel | 184 | V>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9906512 rs754452008 |
186 | E>K | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 187 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1568797471 CA408964324 |
189 | A>G | No |
ClinGen Ensembl |
|
| TCGA novel | 195 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs756494977 CA9906515 |
196 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA9906516 rs370578341 |
199 | R>* | No |
ClinGen ESP ExAC gnomAD |
|
|
CA315204417 rs1000153249 |
202 | G>R | No |
ClinGen TOPMed |
|
|
rs1191599473 CA408964419 |
204 | L>F | No |
ClinGen TOPMed |
|
|
rs1372426323 CA408964426 |
205 | S>G | No |
ClinGen gnomAD |
|
|
rs771341376 CA9906518 |
206 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA408964450 rs1453864113 |
208 | H>R | No |
ClinGen TOPMed |
|
|
rs1224554949 CA408964454 |
209 | G>R | No |
ClinGen gnomAD |
|
|
CA315204445 rs1006731785 |
211 | V>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
| TCGA novel | 220 | G>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 221 | R>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9906524 rs760953373 |
222 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA408964542 rs1234412248 |
222 | S>C | No |
ClinGen gnomAD |
|
|
rs1601415665 CA408964550 |
224 | T>P | No |
ClinGen Ensembl |
|
|
rs1181068233 CA408964567 |
226 | C>Y | No |
ClinGen gnomAD |
|
|
CA408964593 rs1601415670 |
230 | T>P | No |
ClinGen Ensembl |
|
|
CA9906526 rs199565200 |
230 | T>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA408964615 rs763041976 |
233 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9906556 rs572308077 |
236 | D>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1207659580 CA408964642 |
236 | D>N | No |
ClinGen TOPMed |
|
|
rs1311537624 CA408964650 |
237 | K>E | No |
ClinGen gnomAD |
|
|
CA9906559 rs747098306 |
240 | D>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 241 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768787011 CA9906560 |
242 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs1242137245 CA408964699 |
244 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
CA9906563 rs371730106 |
244 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs774164787 CA9906564 |
247 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA408964746 rs1437045880 |
251 | L>S | No |
ClinGen gnomAD |
|
|
CA9906565 rs759459286 |
254 | R>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 256 | F>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA408964792 rs1384590527 |
257 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1452015420 CA408964789 |
257 | R>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 261 | I>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA408964840 rs763558512 |
265 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763558512 CA9906569 |
265 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA408964862 rs1437231621 |
268 | R>C | No |
ClinGen gnomAD |
|
|
rs889277600 CA315204993 |
275 | I>L | No |
ClinGen TOPMed gnomAD |
|
|
CA408964915 rs1467589776 |
276 | E>G | No |
ClinGen gnomAD |
|
|
rs1250785943 CA408964913 |
276 | E>K | No |
ClinGen TOPMed |
|
|
rs1176307007 CA408964929 |
278 | A>V | No |
ClinGen TOPMed |
|
|
CA9906574 rs146919360 |
279 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9906576 rs752071908 CA408964953 |
282 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs967086398 CA315205023 |
284 | D>Y | No |
ClinGen TOPMed |
|
|
rs1363544131 CA408964979 |
285 | S>F | No |
ClinGen gnomAD |
|
| TCGA novel | 286 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs748360940 CA9906581 CA9906580 |
287 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748360940 CA9906579 |
287 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1297475988 CA408965009 |
289 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs752809061 CA9906598 |
290 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408965026 rs1435238590 |
291 | W>G | No |
ClinGen TOPMed |
|
|
rs1328987907 CA408965070 |
297 | E>K | No |
ClinGen TOPMed |
|
|
CA408965081 rs1601416296 |
298 | D>A | No |
ClinGen Ensembl |
|
|
CA9906600 rs778065017 |
299 | L>M | No |
ClinGen ExAC gnomAD |
|
|
rs1410854108 CA408965088 |
299 | L>P | No |
ClinGen TOPMed |
|
|
CA408965098 rs1231034866 |
301 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA315205434 rs749433251 |
302 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749433251 CA408965107 |
302 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9906601 rs749433251 |
302 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs116696243 CA9906603 |
304 | E>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA408965125 rs1251310006 |
305 | H>R | No |
ClinGen gnomAD |
|
|
CA408965123 rs1226045495 |
305 | H>Y | No |
ClinGen gnomAD |
|
|
rs1601416324 CA408965133 |
306 | I>T | No |
ClinGen Ensembl |
|
|
rs1479925061 CA408965141 |
307 | P>L | No |
ClinGen gnomAD |
|
|
CA408965144 rs1182862210 |
308 | P>S | No |
ClinGen TOPMed |
|
|
CA408965153 rs1239562012 |
309 | P>L | No |
ClinGen gnomAD |
|
|
rs747013111 CA9906605 |
310 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs963999479 CA315205478 |
311 | R>Q | No |
ClinGen gnomAD |
|
|
CA9906606 rs369115465 |
311 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1420127286 CA408965172 |
313 | P>L | No |
ClinGen gnomAD |
|
|
rs776371372 CA9906607 |
313 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA408965174 rs1424527605 |
314 | K>E | No |
ClinGen gnomAD |
|
|
rs747697304 CA9906608 |
314 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA9906609 rs769146045 |
315 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs372989529 CA9906610 |
316 | I>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA408965211 rs1568798391 |
320 | H>Y | No |
ClinGen Ensembl |
|
|
CA408965219 rs1325608761 |
321 | N>Y | No |
ClinGen TOPMed |
|
|
rs1374557375 CA408965235 |
323 | K>R | No |
ClinGen gnomAD |
|
| TCGA novel | 324 | C>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs762671904 CA9906611 |
324 | C>R | No |
ClinGen ExAC gnomAD |
|
|
CA9906612 rs766891909 |
324 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs774853431 CA9906613 |
330 | N>D | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 331 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9906614 rs760178512 |
332 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs910605795 CA315205529 |
339 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1329732327 CA408965360 |
340 | E>G | No |
ClinGen TOPMed |
|
|
rs1275800499 CA408965358 |
340 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA9906617 rs377517343 |
341 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1209157184 CA408965384 |
343 | D>V | No |
ClinGen TOPMed gnomAD |
|
|
rs976787239 CA315205530 |
346 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
CA9906618 rs756253013 |
349 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1186030764 CA408965431 |
350 | K>E | No |
ClinGen gnomAD |
|
|
CA9906619 rs764034146 |
350 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA9906620 rs371063445 |
352 | S>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9906622 rs757463233 |
353 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs757463233 CA9906621 |
353 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs751365357 CA9906623 |
355 | N>H | No |
ClinGen ExAC gnomAD |
|
|
rs1432702842 CA408965465 |
355 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs781220884 CA9906626 |
357 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA408965484 rs1363364671 |
358 | P>L | No |
ClinGen gnomAD |
|
|
CA315205566 rs866027737 |
359 | Y>C | No |
ClinGen Ensembl |
|
|
rs777117770 CA9906629 |
360 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1370899418 CA408965499 |
361 | I>F | No |
ClinGen TOPMed gnomAD |
|
|
CA9906632 rs370509709 |
361 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9906630 rs749017764 |
361 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs759986577 CA9906633 COSM1533509 |
362 | E>K | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs759986577 CA408965502 |
362 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778743864 CA9906650 |
365 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA315206480 rs893742215 |
368 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs138343840 CA9906652 |
375 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs777051895 CA9906656 |
376 | G>A | No |
ClinGen ExAC gnomAD |
|
|
CA9906655 rs769052153 |
376 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs144346092 CA9906659 |
378 | S>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1175258966 | 378 | S>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9906658 rs144346092 |
378 | S>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM1027936 rs763052949 CA9906661 |
380 | R>* | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA9906660 rs763052949 |
380 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs752423818 CA9906662 |
380 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9906664 VAR_022013 rs16995304 |
381 | G>S | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| TCGA novel | 381 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9906666 rs146344551 |
382 | A>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1601417203 CA408965945 |
383 | Q>R | No |
ClinGen Ensembl |
|
|
CA315206537 rs778546018 |
386 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs778546018 CA9906667 |
386 | S>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
VAR_022014 rs16995309 CA9906668 |
387 | P>L | associated with low glucose tolerance [UniProt] | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1230084781 CA408966055 |
389 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
COSM1027938 CA315206564 rs984329389 |
392 | P>L | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs1412781369 CA408966137 |
395 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA9906672 rs769020982 |
396 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs139630293 CA9906674 |
398 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs769899825 CA9906675 |
399 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA408966240 rs1568799492 |
400 | D>H | No |
ClinGen Ensembl |
|
|
CA408966261 rs1568799498 |
401 | H>R | No |
ClinGen Ensembl |
|
|
CA408966257 rs1409849933 |
401 | H>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA408966293 rs1400455774 |
404 | S>N | No |
ClinGen gnomAD |
|
| TCGA novel | 410 | L>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1279842769 CA408966399 |
412 | N>D | No |
ClinGen TOPMed gnomAD |
|
|
CA408966421 rs1294562007 |
413 | M>I | No |
ClinGen TOPMed |
|
|
CA9906678 rs766525958 |
413 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA408966429 rs1457853372 |
414 | C>Y | No |
ClinGen TOPMed |
|
|
CA9906680 rs760420089 |
415 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs371791783 CA9906681 |
416 | A>T | No |
ClinGen ESP ExAC gnomAD |
|
|
rs922314694 COSM1412424 CA315206615 |
417 | T>M | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
CA9906682 rs145883911 |
420 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1342682532 CA408966494 |
421 | A>T | No |
ClinGen gnomAD |
|
|
rs549340238 CA9906685 |
422 | G>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs367996563 CA408966503 |
423 | A>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs367996563 CA315206642 |
423 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs367996563 COSM1027940 CA9906687 |
423 | A>T | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs1271273002 CA408966510 |
424 | Y>C | No |
ClinGen TOPMed |
|
|
CA408966516 rs1219900969 |
425 | L>V | No |
ClinGen TOPMed |
|
|
rs946652501 CA315206660 |
428 | R>G | No |
ClinGen TOPMed |
|
|
CA408966539 rs1208059231 |
428 | R>T | No |
ClinGen gnomAD |
|
| TCGA novel | 431 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1003217984 CA315207735 |
432 | N>I | No |
ClinGen TOPMed |
|
|
CA9906718 rs779974899 |
436 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs773071006 CA9906720 |
436 | T>W | No |
ClinGen ExAC gnomAD |
No associated diseases with P18031
4 regional properties for P18031
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Tyrosine-specific protein phosphatase, PTPase domain | 3 - 279 | IPR000242 |
| domain | Tyrosine-specific protein phosphatases domain | 189 - 268 | IPR000387 |
| domain | Protein-tyrosine phosphatase, catalytic | 170 - 276 | IPR003595 |
| active_site | Protein-tyrosine phosphatase, active site | 213 - 223 | IPR016130 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.3.48 | Phosphoric monoester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic side of endoplasmic reticulum membrane | The side (leaflet) of the plasma membrane that faces the cytoplasm. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome | A membrane-bounded organelle that receives incoming material from primary endocytic vesicles that have been generated by clathrin-dependent and clathrin-independent endocytosis; vesicles fuse with the early endosome to deliver cargo for sorting into recycling or degradation pathways. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| mitochondrial crista | Any of the inward folds of the mitochondrial inner membrane. Their number, extent, and shape differ in mitochondria from different tissues and organisms. They appear to be devices for increasing the surface area of the mitochondrial inner membrane, where the enzymes of electron transport and oxidative phosphorylation are found. Their shape can vary with the respiratory state of the mitochondria. |
| mitochondrial matrix | The gel-like material, with considerable fine structure, that lies in the matrix space, or lumen, of a mitochondrion. It contains the enzymes of the tricarboxylic acid cycle and, in some organisms, the enzymes concerned with fatty acid oxidation. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| sorting endosome | A multivesicular body surrounded by and connected with multiple tubular compartments with associated vesicles. |
11 GO annotations of molecular function
| Name | Definition |
|---|---|
| cadherin binding | Binding to cadherin, a type I membrane protein involved in cell adhesion. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| ephrin receptor binding | Binding to an ephrin receptor. |
| insulin receptor binding | Binding to an insulin receptor. |
| non-membrane spanning protein tyrosine phosphatase activity | Catalysis of the reaction: non-membrane spanning protein tyrosine phosphate + H2O = non-membrane spanning protein tyrosine + phosphate. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein phosphatase 2A binding | Binding to protein phosphatase 2A. |
| protein tyrosine phosphatase activity | Catalysis of the reaction: protein tyrosine phosphate + H2O = protein tyrosine + phosphate. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| zinc ion binding | Binding to a zinc ion (Zn). |
26 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton reorganization | A process that is carried out at the cellular level which results in dynamic structural changes to the arrangement of constituent parts of cytoskeletal structures comprising actin filaments and their associated proteins. |
| cellular response to unfolded protein | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an unfolded protein stimulus. |
| endoplasmic reticulum unfolded protein response | The series of molecular signals generated as a consequence of the presence of unfolded proteins in the endoplasmic reticulum (ER) or other ER-related stress; results in changes in the regulation of transcription and translation. |
| growth hormone receptor signaling pathway via JAK-STAT | The process in which STAT proteins (Signal Transducers and Activators of Transcription) are activated by members of the JAK (janus activated kinase) family of tyrosine kinases, following the binding of physiological ligands to the growth hormone receptor. Once activated, STATs dimerize and translocate to the nucleus and modulate the expression of target genes. |
| insulin receptor signaling pathway | The series of molecular signals generated as a consequence of the insulin receptor binding to insulin. |
| IRE1-mediated unfolded protein response | The series of molecular signals mediated by the endoplasmic reticulum stress sensor IRE1 (Inositol-requiring transmembrane kinase/endonuclease). Begins with activation of IRE1 in response to endoplasmic reticulum (ER) stress, and ends with regulation of a downstream cellular process, e.g. transcription. One target of activated IRE1 is the transcription factor HAC1 in yeast, or XBP1 in mammals; IRE1 cleaves an intron of a mRNA coding for HAC1/XBP1 to generate an activated HAC1/XBP1 transcription factor, which controls the up regulation of UPR-related genes. At least in mammals, IRE1 can also signal through additional intracellular pathways including JNK and NF-kappaB. |
| negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of an endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway. |
| negative regulation of ERK1 and ERK2 cascade | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| negative regulation of insulin receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of insulin receptor signaling. |
| negative regulation of MAP kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of MAP kinase activity. |
| negative regulation of PERK-mediated unfolded protein response | Any process that stops, prevents or reduces the frequency, rate or extent of the PERK-mediated unfolded protein response. |
| negative regulation of signal transduction | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction. |
| negative regulation of vascular endothelial growth factor receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of vascular endothelial growth factor receptor signaling pathway activity. |
| peptidyl-tyrosine dephosphorylation | The removal of phosphoric residues from peptidyl-O-phospho-tyrosine to form peptidyl-tyrosine. |
| peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity | Any peptidyl-tyrosine dephosphorylation that is involved in inactivation of protein kinase activity. |
| platelet-derived growth factor receptor-beta signaling pathway | The series of molecular signals initiated by the binding of a ligand to a beta-type platelet-derived growth factor receptor (PDGFbeta) on the surface of a signal-receiving cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| positive regulation of IRE1-mediated unfolded protein response | Any process that activates or increases the frequency, rate or extent of the IRE1-mediated unfolded protein response. |
| positive regulation of JUN kinase activity | Any process that activates or increases the frequency, rate or extent of JUN kinase activity. |
| positive regulation of protein tyrosine kinase activity | Any process that increases the rate, frequency, or extent of protein tyrosine kinase activity. |
| positive regulation of receptor catabolic process | Any process that activates or increases the frequency, rate or extent of receptor catabolic process. |
| protein dephosphorylation | The process of removing one or more phosphoric residues from a protein. |
| regulation of endocytosis | Any process that modulates the frequency, rate or extent of endocytosis. |
| regulation of hepatocyte growth factor receptor signaling pathway | Any process that modulates the frequency, rate or extent of hepatocyte growth factor receptor signaling pathway. |
| regulation of intracellular protein transport | Any process that modulates the frequency, rate or extent of the directed movement of proteins within cells. |
| regulation of signal transduction | Any process that modulates the frequency, rate or extent of signal transduction. |
| regulation of type I interferon-mediated signaling pathway | Any process that modulates the rate, frequency or extent of a type I interferon-mediated signaling pathway. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P17706 | PTPN2 | Tyrosine-protein phosphatase non-receptor type 2 | Homo sapiens (Human) | EV |
| Q06180 | Ptpn2 | Tyrosine-protein phosphatase non-receptor type 2 | Mus musculus (Mouse) | SS |
| P35821 | Ptpn1 | Tyrosine-protein phosphatase non-receptor type 1 | Mus musculus (Mouse) | PR |
| A1L1L3 | Ptpn20 | Tyrosine-protein phosphatase non-receptor type 20 | Rattus norvegicus (Rat) | PR |
| P35233 | Ptpn2 | Tyrosine-protein phosphatase non-receptor type 2 | Rattus norvegicus (Rat) | SS |
| P20417 | Ptpn1 | Tyrosine-protein phosphatase non-receptor type 1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEMEKEFEQI | DKSGSWAAIY | QDIRHEASDF | PCRVAKLPKN | KNRNRYRDVS | PFDHSRIKLH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QEDNDYINAS | LIKMEEAQRS | YILTQGPLPN | TCGHFWEMVW | EQKSRGVVML | NRVMEKGSLK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CAQYWPQKEE | KEMIFEDTNL | KLTLISEDIK | SYYTVRQLEL | ENLTTQETRE | ILHFHYTTWP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DFGVPESPAS | FLNFLFKVRE | SGSLSPEHGP | VVVHCSAGIG | RSGTFCLADT | CLLLMDKRKD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PSSVDIKKVL | LEMRKFRMGL | IQTADQLRFS | YLAVIEGAKF | IMGDSSVQDQ | WKELSHEDLE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PPPEHIPPPP | RPPKRILEPH | NGKCREFFPN | HQWVKEETQE | DKDCPIKEEK | GSPLNAAPYG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| IESMSQDTEV | RSRVVGGSLR | GAQAASPAKG | EPSLPEKDED | HALSYWKPFL | VNMCVATVLT |
| 430 | |||||
| AGAYLCYRFL | FNSNT |