P17676
Gene name |
CEBPB |
Protein name |
CCAAT/enhancer-binding protein beta |
Names |
C/EBP beta, Liver activator protein, LAP, Liver-enriched inhibitory protein, LIP, Nuclear factor NF-IL6, Transcription factor 5, TCF-5 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1051 |
EC number |
|
Protein Class |
CCAAT/ENHANCER BINDING PROTEIN (PTHR23334) |
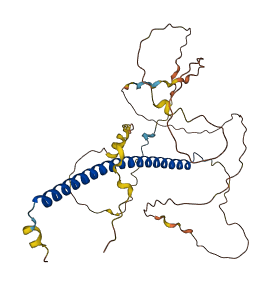
Descriptions
CEBPB is a transcription factor that regulates oncogenic H-RasV12-induced senescence and growth arrest. Three distinct N-terminal sequences that are predicted to form secondary structures inhibit CEBPB DNA binding. These include the autoinhibitory domain, which is located within CR5, and two LX motifs (LX1 and LX2). CEBPB is autoinhibited by these multiple elements, which also play a role in mediating interactions with coactivators. The autoinhibitory region physically interacts with the basic region within the DNA-binding domain to achieve autoinhibition.Truncation of the autoinhibitory domain, LX1, and/or LX2 recovered CEBPB DNA binding activity
Autoinhibitory domains (AIDs)
Target domain |
269-334 (DNA-binding domain (bZIP region)) |
Relief mechanism |
Partner binding, PTM |
Assay |
Deletion assay, Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

16 structures for P17676
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1GTW | X-ray | 185 A | A/B | 259-336 | PDB |
| 1GU4 | X-ray | 180 A | A/B | 259-336 | PDB |
| 1GU5 | X-ray | 210 A | A/B | 259-336 | PDB |
| 1H88 | X-ray | 280 A | A/B | 259-336 | PDB |
| 1H89 | X-ray | 245 A | A/B | 273-336 | PDB |
| 1H8A | X-ray | 223 A | A/B | 259-336 | PDB |
| 1HJB | X-ray | 300 A | A/B/D/E | 259-345 | PDB |
| 1IO4 | X-ray | 300 A | A/B | 259-336 | PDB |
| 2E42 | X-ray | 180 A | A/B | 259-336 | PDB |
| 2E43 | X-ray | 210 A | A/B | 259-336 | PDB |
| 6MG1 | X-ray | 175 A | A/B | 269-344 | PDB |
| 6MG2 | X-ray | 193 A | A/B | 269-344 | PDB |
| 6MG3 | X-ray | 205 A | A/B | 269-344 | PDB |
| 7L4V | X-ray | 175 A | A/B | 269-344 | PDB |
| 7UPZ | X-ray | 249 A | A/B | 257-336 | PDB |
| AF-P17676-F1 | Predicted | AlphaFoldDB |
283 variants for P17676
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA408946977 rs1174067697 |
2 | Q>* | No |
ClinGen gnomAD |
|
|
rs1408002318 CA408947043 |
3 | R>H | No |
ClinGen gnomAD |
|
|
CA9906214 rs751204816 |
7 | W>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1258959413 CA408947154 |
8 | D>N | No |
ClinGen TOPMed |
|
|
CA9906215 rs759182476 |
10 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA408947252 rs1568673101 |
12 | L>P | No |
ClinGen Ensembl |
|
|
rs1326067176 CA408947386 |
17 | P>L | No |
ClinGen gnomAD |
|
|
rs1041584212 CA315203351 |
18 | P>L | No |
ClinGen TOPMed |
|
|
rs1260313083 CA408947391 |
18 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA408947411 rs1206905908 |
19 | P>L | No |
ClinGen gnomAD |
|
|
rs1000008468 CA315203383 |
20 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA9906225 rs377368819 |
20 | A>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA315203405 rs1034526659 |
23 | S>F | No |
ClinGen TOPMed |
|
|
rs746123580 CA9906226 |
24 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA408947547 rs1157927460 |
25 | E>D | No |
ClinGen gnomAD |
|
|
rs1361637653 CA408947563 |
26 | V>A | No |
ClinGen gnomAD |
|
|
CA408947556 rs1422775925 |
26 | V>L | No |
ClinGen gnomAD |
|
|
CA9906227 rs772304270 |
27 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA9906228 rs781028963 |
30 | Y>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1411297267 CA408947648 |
30 | Y>H | No |
ClinGen TOPMed |
|
|
CA408947691 rs1404927663 |
31 | Y>C | No |
ClinGen gnomAD |
|
|
CA408947679 rs747920655 |
31 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747920655 CA9906229 |
31 | Y>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408947708 rs1301797980 |
32 | E>Q | No |
ClinGen gnomAD |
|
|
rs1179373108 CA408947762 |
33 | A>V | No |
ClinGen TOPMed |
|
|
rs769517052 CA9906230 |
34 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs1224443397 CA408947841 |
37 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
CA408947843 rs1224443397 |
37 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1224443397 CA408947840 |
37 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs371495742 CA9906231 |
38 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1241425543 CA408947879 |
39 | A>P | No |
ClinGen gnomAD |
|
|
CA408947888 rs1422705747 |
39 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs770372476 CA9906233 |
41 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408947935 rs770372476 |
41 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9906234 rs773893525 |
42 | G>C | No |
ClinGen ExAC gnomAD |
|
|
CA9906235 rs200084272 |
42 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408947954 rs773893525 |
42 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA315203433 rs200084272 |
42 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9906236 rs767276248 |
43 | K>R | No |
ClinGen ExAC |
|
|
CA315203475 rs867722941 |
44 | A>E | No |
ClinGen Ensembl |
|
|
CA408948045 rs1295266659 |
45 | A>T | No |
ClinGen TOPMed |
|
|
rs1490452851 CA408948072 |
45 | A>V | No |
ClinGen gnomAD |
|
|
CA408948122 rs1176472291 |
47 | A>P | No |
ClinGen gnomAD |
|
|
CA408948158 rs1248138498 |
48 | A>P | No |
ClinGen gnomAD |
|
|
CA315203501 rs753171398 |
49 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs761135515 CA408948190 |
49 | P>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs761135515 CA9906239 |
49 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9906238 rs753171398 |
49 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408948185 rs753171398 |
49 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408948217 rs1384108590 |
50 | P>R | No |
ClinGen gnomAD |
|
|
rs1162239063 CA408948228 |
51 | A>T | No |
ClinGen gnomAD |
|
|
CA408948271 rs1156781585 |
52 | A>V | No |
ClinGen TOPMed |
|
|
CA408948273 rs1437803509 |
53 | R>G | No |
ClinGen gnomAD |
|
|
rs1298012826 CA408948283 |
53 | R>I | No |
ClinGen gnomAD |
|
|
CA315203519 rs1033481854 |
53 | R>S | No |
ClinGen Ensembl |
|
|
rs975638329 CA315203529 |
54 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs975638329 CA315203525 |
54 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs962886579 CA315203521 |
54 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA408948327 rs934133431 |
55 | G>E | No |
ClinGen TOPMed |
|
|
rs1307918746 CA408948306 |
55 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs934133431 CA315203539 |
55 | G>V | No |
ClinGen TOPMed |
|
|
rs533532702 CA315203559 |
56 | P>L | No |
ClinGen 1000Genomes |
|
|
rs896368541 CA315203562 |
57 | R>P | No |
ClinGen Ensembl |
|
|
CA408948401 rs1382138459 |
58 | P>H | No |
ClinGen gnomAD |
|
|
CA408948413 rs987282182 |
59 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA315203572 rs913912949 |
59 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs913912949 CA315203570 |
59 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs987282182 CA315203567 |
59 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1436941998 CA408948508 |
62 | E>G | No |
ClinGen TOPMed |
|
|
CA408948482 rs1249448053 |
62 | E>K | No |
ClinGen gnomAD |
|
|
CA408948531 rs1177452475 |
64 | G>D | No |
ClinGen gnomAD |
|
|
CA408948552 rs1456605960 |
65 | S>R | No |
ClinGen TOPMed |
|
|
CA408948628 rs1601257450 |
67 | G>D | No |
ClinGen Ensembl |
|
|
CA9906243 rs778896468 |
68 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA315203593 rs1055656289 |
69 | H>N | No |
ClinGen TOPMed |
|
|
CA408948781 rs1255238207 |
73 | I>L | No |
ClinGen TOPMed |
|
|
CA315203595 rs551520247 |
77 | P>L | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs1170311066 CA408949027 |
79 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1170311066 CA408949032 |
79 | L>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1224552608 CA408948996 |
79 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1278207940 CA408949042 |
80 | E>Q | No |
ClinGen TOPMed |
|
|
CA9906244 rs750509396 |
81 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1379952079 CA408949216 |
86 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA408949191 rs1317315198 |
86 | Q>K | No |
ClinGen gnomAD |
|
|
CA408949221 rs1434354584 |
87 | A>T | No |
ClinGen TOPMed |
|
|
rs1362562294 CA408949239 |
88 | P>S | No |
ClinGen TOPMed |
|
|
rs1014037357 CA315203602 |
95 | D>A | No |
ClinGen TOPMed gnomAD |
|
|
CA408949390 rs1436160535 |
96 | T>N | No |
ClinGen TOPMed |
|
|
rs1176370107 CA408949375 |
96 | T>P | No |
ClinGen TOPMed |
|
|
CA315203618 rs1024026253 |
97 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1205138242 CA408949418 |
98 | E>Q | No |
ClinGen gnomAD |
|
|
CA408949551 rs1171898938 |
105 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
rs886775109 CA315203637 |
105 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA408949553 rs1171898938 |
105 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1405777201 CA408949560 |
106 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs747170675 CA9906248 |
106 | P>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1405777201 CA408949556 |
106 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA9906249 rs755939066 |
108 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1359843144 CA408949612 |
109 | A>S | No |
ClinGen gnomAD |
|
|
CA408949666 rs1430043245 |
111 | S>F | No |
ClinGen gnomAD |
|
|
CA9906250 rs777498639 |
112 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777498639 CA315203645 |
112 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408949695 rs1312933241 |
113 | Q>H | No |
ClinGen TOPMed |
|
|
rs1377861808 CA408949690 |
113 | Q>P | No |
ClinGen TOPMed |
|
|
CA315203649 rs868698135 |
114 | H>N | No |
ClinGen Ensembl |
|
|
CA408949744 rs1364075169 |
115 | H>R | No |
ClinGen gnomAD |
|
|
CA408949761 rs1262441606 |
116 | D>G | No |
ClinGen gnomAD |
|
|
CA9906255 rs773801449 |
124 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408949976 rs1244778086 |
125 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs745370394 CA9906256 |
125 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1486654464 CA408950001 |
126 | Y>C | No |
ClinGen TOPMed |
|
|
rs1487164082 CA408949992 |
126 | Y>N | No |
ClinGen gnomAD |
|
|
rs775313594 CA9906258 |
127 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA408950030 rs1204640186 |
127 | G>V | No |
ClinGen TOPMed |
|
|
rs760354287 CA9906259 |
128 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs760354287 CA408950051 |
128 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs777022308 CA9906261 |
130 | N>S | No |
ClinGen ExAC TOPMed |
|
|
rs1302415362 CA408950159 |
134 | P>Q | No |
ClinGen gnomAD |
|
|
CA408950161 rs1302415362 |
134 | P>R | No |
ClinGen gnomAD |
|
|
CA315203715 rs865996366 |
134 | P>T | No |
ClinGen Ensembl |
|
|
rs1348029400 CA408950166 |
135 | A>T | No |
ClinGen TOPMed |
|
|
rs1275211281 CA408950230 |
137 | Y>H | No |
ClinGen gnomAD |
|
|
rs1221691260 CA408950265 |
138 | G>A | No |
ClinGen gnomAD |
|
|
rs1349574578 CA408950252 |
138 | G>C | No |
ClinGen gnomAD |
|
|
CA408950247 rs1349574578 |
138 | G>S | No |
ClinGen gnomAD |
|
|
CA9906264 rs750834194 |
139 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs750834194 CA408950269 |
139 | Y>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408950293 rs1353755963 |
140 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA408950297 rs1353755963 |
140 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA408950316 rs1218018800 |
141 | S>C | No |
ClinGen gnomAD |
|
|
CA408950320 rs1268733163 |
141 | S>N | No |
ClinGen gnomAD |
|
|
CA408950346 rs1568673694 |
142 | L>P | No |
ClinGen Ensembl |
|
|
CA408950347 rs1028525078 |
143 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1028525078 CA315203722 |
143 | G>W | No |
ClinGen TOPMed gnomAD |
|
|
rs1157017002 CA408950367 |
144 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA9906267 rs751807082 |
144 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs1157017002 CA408950363 |
144 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
CA315203742 rs868040290 |
145 | L>P | No |
ClinGen Ensembl |
|
|
rs868040290 CA408950383 |
145 | L>Q | No |
ClinGen Ensembl |
|
|
CA408950379 rs1420321062 |
145 | L>V | No |
ClinGen gnomAD |
|
|
CA408950398 rs1162857185 |
146 | G>E | No |
ClinGen gnomAD |
|
|
rs1324018352 CA408950435 |
147 | A>G | No |
ClinGen gnomAD |
|
|
CA408950414 rs777472730 |
147 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9906269 rs777472730 |
147 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408950430 rs1324018352 |
147 | A>V | No |
ClinGen gnomAD |
|
|
CA408950440 rs1280881046 |
148 | A>D | No |
ClinGen gnomAD |
|
|
rs778923544 CA9906272 |
148 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA408950444 rs1280881046 |
148 | A>V | No |
ClinGen gnomAD |
|
|
rs1244522460 CA408950463 |
149 | K>R | No |
ClinGen gnomAD |
|
|
CA9906274 rs771728816 |
151 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA408950505 rs771728816 |
151 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs775117213 CA9906275 |
151 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1234746665 CA408950540 |
153 | H>N | No |
ClinGen gnomAD |
|
|
rs1183129152 CA408950573 |
154 | P>R | No |
ClinGen gnomAD |
|
|
CA408950557 rs1440542889 |
154 | P>T | No |
ClinGen gnomAD |
|
|
CA315203787 rs986907543 |
155 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs552807936 CA408950591 |
155 | G>R | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs552807936 CA315203781 |
155 | G>S | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs1163728431 CA408950632 |
156 | C>Y | No |
ClinGen gnomAD |
|
|
CA408950673 rs1215435519 |
157 | F>L | No |
ClinGen TOPMed |
|
|
rs1407948246 CA408950686 |
158 | A>S | No |
ClinGen gnomAD |
|
|
rs1330481183 CA408950722 |
159 | P>L | No |
ClinGen gnomAD |
|
|
rs1322081722 CA408950711 |
159 | P>T | No |
ClinGen gnomAD |
|
|
rs1021034803 CA315203806 |
161 | H>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1021034803 CA315203791 |
161 | H>P | No |
ClinGen TOPMed gnomAD |
|
|
rs746697753 CA9906276 |
162 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA9906277 rs768417455 |
164 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA315203875 rs935971375 |
165 | P>A | No |
ClinGen TOPMed |
|
|
rs1056106431 CA315203910 |
169 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA408950979 rs1601257957 |
170 | P>A | No |
ClinGen Ensembl |
|
|
rs1179981504 CA408951011 |
171 | A>S | No |
ClinGen Ensembl |
|
|
rs1365763947 CA408951058 |
172 | E>K | No |
ClinGen TOPMed |
|
|
rs1363436869 CA408951068 |
172 | E>V | No |
ClinGen Ensembl |
|
|
rs1568673871 CA408951199 |
177 | P>T | No |
ClinGen Ensembl |
|
|
CA408951228 rs1194501927 |
178 | G>D | No |
ClinGen gnomAD |
|
|
rs949881400 CA315203934 |
181 | P>S | No |
ClinGen TOPMed |
|
|
CA408951342 rs1241391459 |
182 | A>E | No |
ClinGen gnomAD |
|
|
rs1425585166 CA408951393 |
183 | D>V | No |
ClinGen gnomAD |
|
|
rs1188181806 CA408951505 |
187 | K>E | No |
ClinGen gnomAD |
|
|
rs1601258021 CA408951642 |
190 | A>T | No |
ClinGen Ensembl |
|
|
rs1352138512 CA408951704 |
191 | G>R | No |
ClinGen TOPMed |
|
|
rs1232023086 CA408951740 |
192 | A>E | No |
ClinGen TOPMed |
|
|
CA408951724 rs1253901803 |
192 | A>T | No |
ClinGen TOPMed |
|
|
CA315203964 rs886887483 |
193 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs886887483 CA315203990 |
193 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
CA315203957 rs977528485 |
193 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs977528485 CA408951744 |
193 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA9906285 rs763405415 |
194 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763405415 CA315203997 |
194 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
VAR_016300 CA315203998 rs4253440 |
195 | G>S | No |
ClinGen UniProt 1000Genomes TOPMed dbSNP gnomAD |
|
|
CA408951807 rs1396037022 |
195 | G>V | No |
ClinGen gnomAD |
|
|
CA408952006 rs1387171267 |
202 | G>S | No |
ClinGen TOPMed |
|
|
CA408952084 rs1171071138 |
204 | P>R | No |
ClinGen TOPMed |
|
|
rs936053017 CA315204004 |
205 | Y>H | No |
ClinGen Ensembl |
|
|
CA408952130 rs1434311111 |
206 | A>E | No |
ClinGen TOPMed |
|
|
rs1450841958 CA408952113 |
206 | A>S | No |
ClinGen gnomAD |
|
|
CA408952174 rs1395513520 |
208 | R>C | No |
ClinGen TOPMed |
|
|
CA9906286 rs766497042 |
210 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA315204030 rs955586069 |
211 | L>F | No |
ClinGen TOPMed |
|
|
rs1280535006 CA408952406 |
216 | V>L | No |
ClinGen gnomAD |
|
|
rs894690458 CA315204034 |
217 | P>S | No |
ClinGen Ensembl |
|
|
CA408952449 rs1437427883 |
218 | S>G | No |
ClinGen gnomAD |
|
|
rs1177525422 CA408952461 |
218 | S>T | No |
ClinGen gnomAD |
|
|
rs1223154252 CA408952482 |
219 | G>S | No |
ClinGen gnomAD |
|
|
CA408952524 rs1261434142 |
220 | S>N | No |
ClinGen gnomAD |
|
|
CA408952536 rs1021065134 |
221 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
CA315204045 rs1021065134 |
221 | S>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1195720392 CA408952732 |
226 | T>M | No |
ClinGen gnomAD |
|
|
rs1479961355 CA408952794 |
229 | S>L | No |
ClinGen gnomAD |
|
|
rs1173816740 CA408952802 |
230 | S>A | No |
ClinGen gnomAD |
|
|
rs1379073346 CA408952804 |
230 | S>F | No |
ClinGen gnomAD |
|
|
rs1270154591 CA408952842 |
232 | P>Q | No |
ClinGen TOPMed |
|
|
CA408952849 rs1157530393 |
233 | P>S | No |
ClinGen gnomAD |
|
|
rs757080333 CA408952975 |
237 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA9906291 rs757080333 |
237 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA315204076 rs993194118 |
238 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs993194118 CA408952998 |
238 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1281768059 CA408953047 |
240 | D>V | No |
ClinGen gnomAD |
|
|
CA315204100 rs979483213 |
241 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA408953141 rs1234214452 |
243 | A>T | No |
ClinGen gnomAD |
|
|
rs1260263794 CA408953159 |
243 | A>V | No |
ClinGen gnomAD |
|
|
CA408953230 rs1263727087 |
246 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1490163883 CA408953247 |
247 | A>S | No |
ClinGen gnomAD |
|
|
CA408953252 rs1197080663 |
247 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA408953319 rs1269170784 |
250 | A>T | No |
ClinGen gnomAD |
|
|
CA408953332 rs1433450143 |
250 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1361582241 CA408953357 |
251 | G>A | No |
ClinGen gnomAD |
|
|
CA408953381 rs758377322 |
252 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408953368 rs1160981231 |
252 | A>P | No |
ClinGen gnomAD |
|
|
rs758377322 CA9906295 |
252 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs576451769 CA408953401 |
253 | A>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9906296 rs576451769 |
253 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1568674231 CA408953511 |
258 | Q>* | No |
ClinGen Ensembl |
|
|
rs1370147348 CA408953551 |
259 | V>F | No |
ClinGen gnomAD |
|
|
rs1370147348 CA408953548 |
259 | V>I | No |
ClinGen gnomAD |
|
|
rs1370147348 CA408953549 |
259 | V>L | No |
ClinGen gnomAD |
|
|
rs139316476 CA9906298 |
260 | K>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1387599771 CA408953571 |
260 | K>R | No |
ClinGen gnomAD |
|
|
rs1334074288 CA408953593 |
261 | S>G | No |
ClinGen gnomAD |
|
|
CA9906299 rs780897586 |
261 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs747712410 CA9906300 |
263 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs770219852 CA9906301 |
264 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs763493442 CA9906303 |
267 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9906302 rs773431251 |
267 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408953754 rs1568674270 |
269 | K>Q | No |
ClinGen Ensembl |
|
|
rs774473666 CA9906305 |
271 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs949986667 CA315204176 |
272 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1352010258 CA408953825 |
273 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1352010258 CA408953827 |
273 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1163247707 CA408953822 |
273 | E>G | No |
ClinGen gnomAD |
|
|
rs1444609103 CA408953816 |
273 | E>Q | No |
ClinGen gnomAD |
|
|
rs1297957411 CA408953858 |
275 | K>M | No |
ClinGen gnomAD |
|
|
rs1449245373 CA408953883 |
277 | R>Q | No |
ClinGen gnomAD |
|
|
rs1398446653 CA408953882 |
277 | R>W | No |
ClinGen gnomAD |
|
|
rs1249391818 CA408954103 |
289 | R>H | No |
ClinGen gnomAD |
|
|
CA408954125 rs1198850318 |
290 | D>G | No |
ClinGen gnomAD |
|
|
rs1459556014 CA408955116 |
292 | A>T | No |
ClinGen gnomAD |
|
|
rs750238105 COSM1412416 CA408955153 |
293 | K>N | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA408955169 rs1416508856 |
294 | M>I | No |
ClinGen gnomAD |
|
|
rs868421494 CA315204205 |
295 | R>C | No |
ClinGen Ensembl |
|
|
rs1179622728 CA408955245 |
298 | E>A | No |
ClinGen TOPMed |
|
|
rs1184743924 CA408955351 |
302 | K>N | No |
ClinGen gnomAD |
|
|
CA408955344 rs1405457475 |
302 | K>R | No |
ClinGen gnomAD |
|
|
rs1213898726 CA408955436 |
306 | L>F | No |
ClinGen gnomAD |
|
|
rs1218889971 CA408955535 |
311 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA9906319 rs778180638 |
318 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs749640653 CA9906320 |
319 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA408955748 rs771228331 |
320 | L>M | No |
ClinGen ExAC gnomAD |
|
|
rs1478974180 CA408955838 |
325 | S>G | No |
ClinGen gnomAD |
|
|
CA315204281 rs867148328 |
328 | R>W | No |
ClinGen Ensembl |
|
|
rs1178388370 CA408955905 |
330 | L>F | No |
ClinGen gnomAD |
|
|
CA9906326 rs761024259 |
332 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1342163109 CA408955984 |
336 | E>D | No |
ClinGen gnomAD |
|
|
rs1342163109 CA408955986 |
336 | E>D | No |
ClinGen gnomAD |
|
|
rs1601258595 CA408955977 |
336 | E>Q | No |
ClinGen Ensembl |
|
|
CA408955990 rs1400314604 |
337 | P>A | No |
ClinGen gnomAD |
|
|
rs1400314604 CA408955988 |
337 | P>T | No |
ClinGen gnomAD |
|
|
CA9906329 rs762842653 |
339 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs766233869 CA9906330 |
340 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA408956024 rs766233869 |
340 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751561149 CA9906331 |
341 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA408956037 rs751561149 |
341 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA408956041 rs1315756377 |
341 | S>Y | No |
ClinGen gnomAD |
No associated diseases with P17676
1 regional properties for P17676
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Basic-leucine zipper domain | 269 - 334 | IPR004827 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR23334 | CCAAT/ENHANCER BINDING PROTEIN |
| PANTHER Subfamily | PTHR23334:SF21 | CCAAT_ENHANCER-BINDING PROTEIN BETA |
| PANTHER Protein Class |
DNA-binding transcription factor
basic leucine zipper transcription factor |
|
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| C/EBP complex | A dimeric, sequence specific DNA-binding transcription factor complex regulating the expression of genes involved in immune and inflammatory responses. Exists at least as alpha and beta homodimeric forms. Binds to regulatory regions of several acute-phase and cytokines genes and probably plays a role in the regulation of acute-phase reaction, inflammation and hemopoiesis. The consensus recognition site is 5'-T |
| CHOP-C/EBP complex | A heterodimeric protein complex that is composed of the transcription factor CHOP (GADD153) and a member of the C/EBP family of transcription factors. |
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| condensed chromosome, centromeric region | The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nuclear matrix | The dense fibrillar network lying on the inner side of the nuclear membrane. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| RNA polymerase II transcription regulator complex | A transcription factor complex that acts at a regulatory region of a gene transcribed by RNA polymerase II. |
18 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription repressor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that represses or decreases the transcription of specific gene sets transcribed by RNA polymerase II. |
| glucocorticoid receptor binding | Binding to a nuclear glucocorticoid receptor. |
| histone acetyltransferase binding | Binding to an histone acetyltransferase. |
| histone deacetylase binding | Binding to histone deacetylase. |
| kinase binding | Binding to a kinase, any enzyme that catalyzes the transfer of a phosphate group. |
| protein heterodimerization activity | Binding to a nonidentical protein to form a heterodimer. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| RNA polymerase II core promoter sequence-specific DNA binding | Binding to a DNA sequence that is part of the core promoter of a RNA polymerase II-transcribed gene. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
| ubiquitin-like protein ligase binding | Binding to a ubiquitin-like protein ligase, such as ubiquitin-ligase. |
41 GO annotations of biological process
| Name | Definition |
|---|---|
| acute-phase response | An acute inflammatory response that involves non-antibody proteins whose concentrations in the plasma increase in response to infection or injury of homeothermic animals. |
| brown fat cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a brown adipocyte, an animal connective tissue cell involved in adaptive thermogenesis. Brown adipocytes contain multiple small droplets of triglycerides and a high number of mitochondria. |
| cellular response to amino acid stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an amino acid stimulus. An amino acid is a carboxylic acids containing one or more amino groups. |
| cellular response to interleukin-1 | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-1 stimulus. |
| cellular response to lipopolysaccharide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| cellular response to organic cyclic compound | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic cyclic compound stimulus. |
| defense response to bacterium | Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism. |
| embryonic placenta development | The embryonically driven process whose specific outcome is the progression of the placenta over time, from its formation to the mature structure. The placenta is an organ of metabolic interchange between fetus and mother, partly of embryonic origin and partly of maternal origin. |
| granuloma formation | The formation of nodular inflammatory lesions, usually small or granular, firm, persistent, well-structured, and containing compactly grouped T lymphocytes and modified phagocytes such as epithelioid cells, giant cells, and other macrophages. Granuloma formation represents a chronic inflammatory response initiated by various infectious and noninfectious agents. The center of a granuloma consists of fused macrophages, which can become necrotic. |
| hepatocyte proliferation | The multiplication or reproduction of hepatocytes, resulting in the expansion of a cell population. Hepatocytes form the main structural component of the liver. They are specialized epithelial cells that are organized into interconnected plates called lobules. |
| immune response | Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat. |
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | The series of molecular signals in which an intracellular signal is conveyed to trigger the apoptotic death of a cell. The pathway is induced in response to a stimulus indicating endoplasmic reticulum (ER) stress, and ends when the execution phase of apoptosis is triggered. ER stress usually results from the accumulation of unfolded or misfolded proteins in the ER lumen. |
| liver regeneration | The regrowth of lost or destroyed liver. |
| mammary gland epithelial cell differentiation | The process in which a relatively unspecialized epithelial cell becomes a more specialized epithelial cell of the mammary gland. |
| mammary gland epithelial cell proliferation | The multiplication or reproduction of mammary gland epithelial cells, resulting in the expansion of a cell population. Mammary gland epithelial cells make up the covering of surfaces of the mammary gland. The mammary gland is a large compound sebaceous gland that in female mammals is modified to secrete milk. |
| memory | The activities involved in the mental information processing system that receives (registers), modifies, stores, and retrieves informational stimuli. The main stages involved in the formation and retrieval of memory are encoding (processing of received information by acquisition), storage (building a permanent record of received information as a result of consolidation) and retrieval (calling back the stored information and use it in a suitable way to execute a given task). |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| negative regulation of T cell proliferation | Any process that stops, prevents or reduces the rate or extent of T cell proliferation. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| neuron differentiation | The process in which a relatively unspecialized cell acquires specialized features of a neuron. |
| ovarian follicle development | The process whose specific outcome is the progression of the ovarian follicle over time, from its formation to the mature structure. |
| positive regulation of biomineral tissue development | Any process that activates or increases the frequency, rate or extent of biomineral tissue development, the formation of hard tissues that consist mainly of inorganic compounds. |
| positive regulation of cold-induced thermogenesis | Any process that activates or increases the frequency, rate or extent of cold-induced thermogenesis. |
| positive regulation of fat cell differentiation | Any process that activates or increases the frequency, rate or extent of adipocyte differentiation. |
| positive regulation of inflammatory response | Any process that activates or increases the frequency, rate or extent of the inflammatory response. |
| positive regulation of interleukin-4 production | Any process that activates or increases the frequency, rate, or extent of interleukin-4 production. |
| positive regulation of osteoblast differentiation | Any process that activates or increases the frequency, rate or extent of osteoblast differentiation. |
| positive regulation of sodium-dependent phosphate transport | Any process that activates or increases the frequency, rate or extent of sodium-dependent phosphate transport. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter as a result of an endoplasmic reticulum stress. |
| positive regulation of transcription, DNA-templated | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| regulation of cell differentiation | Any process that modulates the frequency, rate or extent of cell differentiation, the process in which relatively unspecialized cells acquire specialized structural and functional features. |
| regulation of dendritic cell differentiation | Any process that modulates the frequency, rate or extent of dendritic cell differentiation. |
| regulation of interleukin-6 production | Any process that modulates the frequency, rate, or extent of interleukin-6 production. |
| regulation of odontoblast differentiation | Any process that modulates the frequency, rate or extent of odontoblast differentiation. |
| regulation of osteoclast differentiation | Any process that modulates the frequency, rate or extent of osteoclast differentiation. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| regulation of transcription, DNA-templated | Any process that modulates the frequency, rate or extent of cellular DNA-templated transcription. |
| response to endoplasmic reticulum stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stress acting at the endoplasmic reticulum. ER stress usually results from the accumulation of unfolded or misfolded proteins in the ER lumen. |
| T-helper 1 cell activation | The change in morphology and behavior of a T-helper 1 cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O02755 | CEBPB | CCAAT/enhancer-binding protein beta | Bos taurus (Bovine) | SS |
| Q90Z72 | NFIL3 | Nuclear factor interleukin-3-regulated protein | Gallus gallus (Chicken) | PR |
| Q16649 | NFIL3 | Nuclear factor interleukin-3-regulated protein | Homo sapiens (Human) | PR |
| O08750 | Nfil3 | Nuclear factor interleukin-3-regulated protein | Mus musculus (Mouse) | PR |
| P28033 | Cebpb | CCAAT/enhancer-binding protein beta | Mus musculus (Mouse) | SS |
| Q6IMZ0 | Nfil3 | Nuclear factor interleukin-3-regulated protein | Rattus norvegicus (Rat) | PR |
| P21272 | Cebpb | CCAAT/enhancer-binding protein beta | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQRLVAWDPA | CLPLPPPPPA | FKSMEVANFY | YEADCLAAAY | GGKAAPAAPP | AARPGPRPPA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GELGSIGDHE | RAIDFSPYLE | PLGAPQAPAP | ATATDTFEAA | PPAPAPAPAS | SGQHHDFLSD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LFSDDYGGKN | CKKPAEYGYV | SLGRLGAAKG | ALHPGCFAPL | HPPPPPPPPP | AELKAEPGFE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PADCKRKEEA | GAPGGGAGMA | AGFPYALRAY | LGYQAVPSGS | SGSLSTSSSS | SPPGTPSPAD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AKAPPTACYA | GAAPAPSQVK | SKAKKTVDKH | SDEYKIRRER | NNIAVRKSRD | KAKMRNLETQ |
| 310 | 320 | 330 | 340 | ||
| HKVLELTAEN | ERLQKKVEQL | SRELSTLRNL | FKQLPEPLLA | SSGHC |