P17210
Gene name |
Khc (kin) |
Protein name |
Kinesin heavy chain |
Names |
|
Species |
Drosophila melanogaster (Fruit fly) |
KEGG Pathway |
dme:Dmel_CG7765 |
EC number |
|
Protein Class |
CENTROMERE PROTEIN E (PTHR47968) |
Descriptions
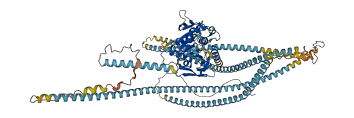
Kinesin-1 is a plus-end microtubule-based motor, and defects in kinesin-based transport are associated with diseases including neurodegeneration.Kinesin is autoinhibited through head-tail interactions. The autoinhibition is released upon cargo recruitment. In Drosophila, centrioles are transported along MTs by Kinesin-1 via direct interaction of the kinesin heavy chain (KHC) with the centriolar protein Pericentrin-like-protein (PLP).
Autoinhibitory domains (AIDs)
Target domain |
10-341 (Kinesin motor domain) |
Relief mechanism |
Partner binding |
Assay |
Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
References
- Kaan HY et al. (2011) "The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition", Science (New York, N.Y.), 333, 883-5
- Korten T et al. (2018) "An automated in vitro motility assay for high-throughput studies of molecular motors", Lab on a chip, 18, 3196-3206
- Hannaford MR et al. (2022) "Pericentrin interacts with Kinesin-1 to drive centriole motility", The Journal of cell biology, 221,
Autoinhibited structure
Activated structure
7 structures for P17210
No variants for P17210
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P17210 | |||||
No associated diseases with P17210
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47968 | CENTROMERE PROTEIN E |
| PANTHER Subfamily | PTHR47968:SF73 | KINESIN HEAVY CHAIN ISOFORM X1 |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cap | Polarized accumulation of cytoskeletal proteins (including F-actin) and regulatory proteins in a cell. An example of this is the actin cap found in Saccharomyces cerevisiae. |
| axon cytoplasm | Any cytoplasm that is part of a axon. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendrite cytoplasm | All of the contents of a dendrite, excluding the surrounding plasma membrane. |
| kinesin complex | Any complex that includes a dimer of molecules from the kinesin superfamily, a group of related proteins that contain an extended region of predicted alpha-helical coiled coil in the main chain that likely produces dimerization. The native complexes of several kinesin family members have also been shown to contain additional peptides, often designated light chains as all of the noncatalytic subunits that are currently known are smaller than the chain that contains the motor unit. Kinesin complexes generally possess a force-generating enzymatic activity, or motor, which converts the free energy of the gamma phosphate bond of ATP into mechanical work. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| microtubule plus-end | The growing (plus) end of a microtubule. In vitro, microtubules polymerize more quickly at the plus end than at the minus end. In vivo, microtubule growth occurs only at the plus end, and the plus end switches between periods of growth and shortening, a behavior known as dynamic instability. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| cytoskeletal motor activity | Generation of force resulting in movement, for example along a microfilament or microtubule, or in torque resulting in membrane scission or rotation of a flagellum. The energy required is obtained either from the hydrolysis of a nucleoside triphosphate or by an electrochemical proton gradient (proton-motive force). |
| microtubule binding | Binding to a microtubule, a filament composed of tubulin monomers. |
| microtubule motor activity | A motor activity that generates movement along a microtubule, driven by ATP hydrolysis. |
| plus-end-directed microtubule motor activity | A motor activity that generates movement along a microtubule toward the plus end, driven by ATP hydrolysis. |
| tropomyosin binding | Binding to tropomyosin, a protein associated with actin filaments both in cytoplasm and, in association with troponin, in the thin filament of striated muscle. |
29 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament bundle organization | A process that results in the assembly, arrangement of constituent parts, or disassembly of an actin filament bundle. |
| anterograde axonal transport of mitochondrion | The directed movement of mitochondria along microtubules in axons away from the cell body and towards the presynapse. |
| anterograde dendritic transport | The directed movement of organelles or molecules along microtubules from the cell body toward the postsynapse in dendrites. |
| anterograde dendritic transport of neurotransmitter receptor complex | The directed movement of a neurotransmitter receptor complex along microtubules in nerve cell dendrites towards the postsynapse. |
| axo-dendritic transport | The directed movement of organelles or molecules along microtubules in neuron projections. |
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| axonogenesis | De novo generation of a long process of a neuron, including the terminal branched region. Refers to the morphogenesis or creation of shape or form of the developing axon, which carries efferent (outgoing) action potentials from the cell body towards target cells. |
| centrosome separation | The process in which duplicated centrosome components move away from each other. The centriole pair within each centrosome becomes part of a separate microtubule organizing center that nucleates a radial array of microtubules called an aster. The two asters move to opposite sides of the nucleus to form the two poles of the mitotic spindle. |
| cytoplasmic transport, nurse cell to oocyte | The directed movement of cytoplasmic constituents synthesized in the nurse cells to the oocyte. |
| cytoskeleton-dependent intracellular transport | The directed movement of substances along cytoskeletal fibers such as microfilaments or microtubules within a cell. |
| dendrite morphogenesis | The process in which the anatomical structures of a dendrite are generated and organized. |
| dorsal appendage formation | Establishment of the dorsal filaments, elaborate specializations of the chorion that protrude from the anterior end of the egg and facilitate embryonic respiration. |
| eye photoreceptor cell differentiation | The process in which a relatively unspecialized cell acquires the specialized features of a photoreceptor cell, as found in the eye, the primary visual organ of most organisms. |
| intracellular distribution of mitochondria | Any process that establishes the spatial arrangement of mitochondria within the cell. |
| larval locomotory behavior | Locomotory behavior in a larval (immature) organism. |
| larval somatic muscle development | The process whose specific outcome is the progression of the larval somatic muscle over time, from its formation to the mature structure. |
| microtubule polymerization | The addition of tubulin heterodimers to one or both ends of a microtubule. |
| microtubule sliding | The movement of one microtubule along another microtubule. |
| microtubule-based movement | A microtubule-based process that results in the movement of organelles, other microtubules, or other cellular components. Examples include motor-driven movement along microtubules and movement driven by polymerization or depolymerization of microtubules. |
| mitochondrion distribution | Any process that establishes the spatial arrangement of mitochondria between and within cells. |
| nuclear migration | The directed movement of the nucleus to a specific location within a cell. |
| oocyte dorsal/ventral axis specification | The establishment, maintenance and elaboration of the dorsal/ventral axis of the oocyte. An example of this is found in Drosophila melanogaster. |
| oocyte microtubule cytoskeleton polarization | Establishment and maintenance of a specific axis of polarity of the oocyte microtubule network. The axis is set so that the minus and plus ends of the microtubules of the mid stage oocyte are positioned along the anterior cortex and at the posterior pole, respectively. An example of this is found in Drosophila melanogaster. |
| pole plasm assembly | Establishment of the specialized cytoplasm found at the poles of the egg. An example of this is found in Drosophila melanogaster. |
| pole plasm oskar mRNA localization | Any process in which oskar mRNA is transported to, or maintained in, the oocyte pole plasm. |
| regulation of pole plasm oskar mRNA localization | Any process that modulates the frequency, rate or extent of the process in which oskar mRNA is transported to, or maintained in, the oocyte pole plasm. |
| stress granule disassembly | The disaggregation of a stress granule into its constituent protein and RNA parts. |
| synaptic vesicle transport | The directed movement of synaptic vesicles. |
| transport along microtubule | The movement of organelles or other particles from one location in the cell to another along microtubules, driven by motor activity. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P33176 | KIF5B | Kinesin-1 heavy chain | Homo sapiens (Human) | EV |
| Q12840 | KIF5A | Kinesin heavy chain isoform 5A | Homo sapiens (Human) | EV |
| O60282 | KIF5C | Kinesin heavy chain isoform 5C | Homo sapiens (Human) | EV |
| P28738 | Kif5c | Kinesin heavy chain isoform 5C | Mus musculus (Mouse) | SS |
| P33175 | Kif5a | Kinesin heavy chain isoform 5A | Mus musculus (Mouse) | SS |
| Q61768 | Kif5b | Kinesin-1 heavy chain | Mus musculus (Mouse) | EV |
| Q2PQA9 | Kif5b | Kinesin-1 heavy chain | Rattus norvegicus (Rat) | SS |
| Q6QLM7 | Kif5a | Kinesin heavy chain isoform 5A | Rattus norvegicus (Rat) | SS |
| P34540 | unc-116 | Kinesin heavy chain | Caenorhabditis elegans | SS |
| Q9SV36 | KINUC | Kinesin-like protein KIN-UC | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSAEREIPAE | DSIKVVCRFR | PLNDSEEKAG | SKFVVKFPNN | VEENCISIAG | KVYLFDKVFK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PNASQEKVYN | EAAKSIVTDV | LAGYNGTIFA | YGQTSSGKTH | TMEGVIGDSV | KQGIIPRIVN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DIFNHIYAME | VNLEFHIKVS | YYEIYMDKIR | DLLDVSKVNL | SVHEDKNRVP | YVKGATERFV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SSPEDVFEVI | EEGKSNRHIA | VTNMNEHSSR | SHSVFLINVK | QENLENQKKL | SGKLYLVDLA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GSEKVSKTGA | EGTVLDEAKN | INKSLSALGN | VISALADGNK | THIPYRDSKL | TRILQESLGG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NARTTIVICC | SPASFNESET | KSTLDFGRRA | KTVKNVVCVN | EELTAEEWKR | RYEKEKEKNA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RLKGKVEKLE | IELARWRAGE | TVKAEEQINM | EDLMEASTPN | LEVEAAQTAA | AEAALAAQRT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ALANMSASVA | VNEQARLATE | CERLYQQLDD | KDEEINQQSQ | YAEQLKEQVM | EQEELIANAR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| REYETLQSEM | ARIQQENESA | KEEVKEVLQA | LEELAVNYDQ | KSQEIDNKNK | DIDALNEELQ |
| 550 | 560 | 570 | 580 | 590 | 600 |
| QKQSVFNAAS | TELQQLKDMS | SHQKKRITEM | LTNLLRDLGE | VGQAIAPGES | SIDLKMSALA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GTDASKVEED | FTMARLFISK | MKTEAKNIAQ | RCSNMETQQA | DSNKKISEYE | KDLGEYRLLI |
| 670 | 680 | 690 | 700 | 710 | 720 |
| SQHEARMKSL | QESMREAENK | KRTLEEQIDS | LREECAKLKA | AEHVSAVNAE | EKQRAEELRS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| MFDSQMDELR | EAHTRQVSEL | RDEIAAKQHE | MDEMKDVHQK | LLLAHQQMTA | DYEKVRQEDA |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EKSSELQNII | LTNERREQAR | KDLKGLEDTV | AKELQTLHNL | RKLFVQDLQQ | RIRKNVVNEE |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SEEDGGSLAQ | KQKISFLENN | LDQLTKVHKQ | LVRDNADLRC | ELPKLEKRLR | CTMERVKALE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| TALKEAKEGA | MRDRKRYQYE | VDRIKEAVRQ | KHLGRRGPQA | QIAKPIRSGQ | GAIAIRGGGA |
| 970 | |||||
| VGGPSPLAQV | NPVNS |