P16333
Gene name |
NCK1 (NCK) |
Protein name |
Cytoplasmic protein NCK1 |
Names |
NCK adaptor protein 1, Nck-1, SH2/SH3 adaptor protein NCK-alpha |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4690 |
EC number |
|
Protein Class |
|
Descriptions
(Annotation based on sequence homology with O43639)
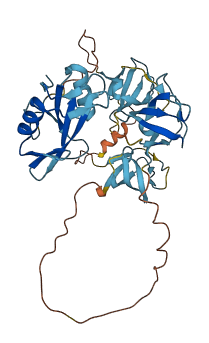
NCK2 is a cytoplasmic protein participating in the reorganization of the actin cytoskeleton and in the formation of the immunological synapses. The autoinhibitory domain locates at a conserved non-canonical (K/R)x(K/R)RxxS sequence, specifically at residues 60-110 linker between the SH3.1 and SH3.2. The autoinhibitory domain binds to the SH3.2 domain via acidic electrostatic potential on the SH3 domain surface and masks the proline-rich sequence (PRS) binding site of SH3.2 from its ligands. Only high affinity stable interactions can overcome the inhibitory interaction; such as phosphorylation of Ser85 at the interaction site.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

16 structures for P16333
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2CI8 | X-ray | 180 A | A | 281-377 | PDB |
| 2CI9 | X-ray | 150 A | A/B | 281-377 | PDB |
| 2CUB | NMR | - | A | 99-173 | PDB |
| 2JS0 | NMR | - | A | 107-165 | PDB |
| 2JS2 | NMR | - | A | 1-61 | PDB |
| 2JW4 | NMR | - | A | 1-63 | PDB |
| 5QU1 | X-ray | 108 A | A/B | 4-59 | PDB |
| 5QU2 | X-ray | 104 A | A/B | 1-59 | PDB |
| 5QU3 | X-ray | 102 A | A/B | 4-59 | PDB |
| 5QU4 | X-ray | 105 A | A/B/C/D | 1-61 | PDB |
| 5QU5 | X-ray | 111 A | A/B | 1-61 | PDB |
| 5QU6 | X-ray | 182 A | 1/2/A/B/C/D/E/F/G/H/I/J/K/L/M/N/O/P/Q/R/S/T/U/V/W/X/Y/Z | 1-61 | PDB |
| 5QU7 | X-ray | 127 A | A/B | 4-59 | PDB |
| 5QU8 | X-ray | 093 A | A | 1-61 | PDB |
| 5QUA | X-ray | 151 A | A/B | 1-61 | PDB |
| AF-P16333-F1 | Predicted | AlphaFoldDB |
244 variants for P16333
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 2 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1616967 rs767008989 CA2633554 |
4 | E>V | liver [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA2633556 rs374161071 |
5 | V>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1486789847 CA354745543 |
5 | V>L | No |
ClinGen gnomAD |
|
| TCGA novel | 5 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs548741298 CA2633557 |
7 | V>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA84315242 rs939753298 |
8 | V>A | No |
ClinGen TOPMed |
|
|
CA354745567 rs1474600867 |
9 | A>G | No |
ClinGen gnomAD |
|
|
rs933930959 CA84315243 |
10 | K>T | No |
ClinGen Ensembl |
|
|
CA354745608 rs1315477618 |
15 | A>S | No |
ClinGen TOPMed |
|
|
CA354745642 rs1340314926 |
19 | Q>H | No |
ClinGen TOPMed |
|
|
CA2633562 rs750326518 |
22 | D>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 27 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA354745708 rs1297134708 |
28 | R>T | No |
ClinGen gnomAD |
|
|
CA354745721 rs1432713512 |
30 | W>* | No |
ClinGen gnomAD |
|
|
CA2633564 rs780159643 |
30 | W>R | No |
ClinGen ExAC gnomAD |
|
|
rs1344285804 CA354745731 |
31 | L>P | No |
ClinGen gnomAD |
|
|
rs781322778 CA2633567 |
33 | D>G | No |
ClinGen ExAC |
|
|
CA2633568 rs748380862 |
37 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1211887220 CA354745775 |
38 | W>L | No |
ClinGen gnomAD |
|
|
CA354745784 rs1278916013 |
39 | W>* | No |
ClinGen gnomAD |
|
|
CA2633569 rs778198006 COSM728221 |
40 | R>* | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs773593965 CA2633570 |
40 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1195839831 CA354745801 COSM1038929 |
42 | R>Q | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA354745819 rs1243005880 |
45 | M>V | No |
ClinGen gnomAD |
|
|
CA2633572 rs771394511 |
51 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA2633573 rs775056529 |
53 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs1167014161 CA354745882 |
54 | N>T | No |
ClinGen TOPMed |
|
|
rs1402063791 CA354745928 |
60 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA354745927 rs1402063791 |
60 | N>T | No |
ClinGen TOPMed gnomAD |
|
|
rs761581267 CA2633577 |
61 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs148772850 CA2633578 |
62 | A>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs537275699 CA2633580 |
63 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs537275699 CA2633579 |
63 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA354745945 COSM84829 rs1438631551 |
63 | R>W | pancreas [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA2633581 rs779922033 |
64 | K>E | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 67 | I>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2633583 rs771451936 |
67 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2633584 rs370617853 |
68 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA354746003 rs1285395879 |
72 | K>R | No |
ClinGen gnomAD |
|
|
rs770040215 CA2633586 |
73 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2633609 rs779358036 |
78 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA354652996 rs1453551155 |
79 | K>I | No |
ClinGen TOPMed |
|
|
CA2633610 rs746338229 |
80 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA354653024 rs1250697408 |
83 | K>I | No |
ClinGen gnomAD |
|
| TCGA novel | 84 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA354653027 rs1184323958 |
84 | P>S | No |
ClinGen TOPMed |
|
|
rs183326283 CA2633612 |
85 | S>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA354653067 rs1374887016 |
87 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA2633613 rs747727219 |
88 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1314939238 CA354653099 |
90 | A>P | No |
ClinGen gnomAD |
|
|
CA2633614 rs769432770 |
91 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs772680921 CA2633615 |
92 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs185485335 CA2633618 |
93 | A>G | No |
ClinGen 1000Genomes ExAC |
|
|
rs762687588 CA2633616 |
93 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA354653171 rs1349759817 |
94 | D>G | No |
ClinGen TOPMed |
|
|
rs774239593 CA2633619 |
96 | S>I | No |
ClinGen ExAC gnomAD |
|
|
CA83833757 rs759443661 |
98 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs759443661 CA354653233 |
98 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA2633620 rs759443661 |
98 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs767504687 CA2633621 |
101 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs752774458 CA2633622 |
102 | E>* | No |
ClinGen ExAC gnomAD |
|
|
rs1257866698 CA354653296 |
102 | E>G | No |
ClinGen gnomAD |
|
|
rs773344757 CA83833772 |
103 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA2633623 rs756123065 |
103 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA354653310 rs1263524026 |
104 | L>F | No |
ClinGen gnomAD |
|
| TCGA novel | 104 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1198000788 CA354653326 |
106 | D>G | No |
ClinGen gnomAD |
|
|
CA83833783 rs1029293238 |
108 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs757532603 CA2633626 |
110 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757532603 CA83833791 |
110 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1056277981 CA83833803 |
111 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA2633628 rs368340310 |
112 | Y>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs141442402 CA83833815 |
116 | N>D | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs780640959 CA2633630 |
117 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA2633632 rs769135774 |
118 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs531554309 CA2633635 |
126 | S>* | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs531554309 CA2633636 |
126 | S>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs748857974 CA2633634 |
126 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA2633637 rs147481903 |
128 | I>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 130 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2633638 rs771863858 |
130 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs775351154 CA2633639 |
131 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1191017779 CA354653695 |
131 | T>I | No |
ClinGen TOPMed |
|
|
CA354653705 rs1486039281 |
133 | V>L | No |
ClinGen gnomAD |
|
|
CA2633640 rs760631298 |
134 | I>F | No |
ClinGen ExAC gnomAD |
|
|
CA354653715 rs764042177 |
134 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1195632737 CA354653727 |
136 | M>I | No |
ClinGen gnomAD |
|
|
CA2633642 rs140020695 |
136 | M>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA354653730 rs1425905946 |
137 | E>K | No |
ClinGen gnomAD |
|
|
rs1418756566 CA354653762 |
141 | D>N | No |
ClinGen gnomAD |
|
|
rs762533477 CA2633643 |
145 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA354653796 rs1347165787 |
145 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1347165787 CA354653798 |
145 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1007197278 CA83833883 |
146 | G>D | No |
ClinGen gnomAD |
|
| TCGA novel | 149 | N>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1336387776 CA354653823 |
149 | N>S | No |
ClinGen TOPMed |
|
|
rs765553907 CA2633644 |
154 | W>C | No |
ClinGen ExAC gnomAD |
|
|
CA83833892 rs11547830 |
156 | P>H | No |
ClinGen Ensembl |
|
|
CA83833898 rs1039212149 |
159 | Y>C | No |
ClinGen Ensembl |
|
|
rs1303546100 CA354653907 |
161 | T>S | No |
ClinGen gnomAD |
|
|
rs780585889 CA2633647 |
162 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1221730094 CA354653921 |
163 | E>D | No |
ClinGen gnomAD |
|
|
rs768735016 CA2633649 |
166 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA354653942 rs1212882193 |
166 | S>R | No |
ClinGen gnomAD |
|
|
CA2633650 rs777123597 |
167 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA2633653 rs778474804 |
171 | H>L | No |
ClinGen ExAC gnomAD |
|
|
CA83833916 rs11547829 |
171 | H>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs778474804 CA354653972 |
171 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA2633652 rs11547829 |
171 | H>Y | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs998315077 CA83833919 |
172 | V>M | No |
ClinGen gnomAD |
|
|
rs746766223 CA83833922 |
173 | G>C | No |
ClinGen Ensembl |
|
|
CA83833930 rs981473337 |
176 | S>T | No |
ClinGen TOPMed |
|
|
VAR_051228 rs13320485 CA2633656 |
180 | A>V | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs149750260 CA2633657 |
183 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs145545353 CA354654050 |
184 | N>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs145545353 COSM1495364 CA2633658 |
184 | N>S | kidney [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA354654067 rs1299355361 |
187 | N>D | No |
ClinGen gnomAD |
|
|
rs765244758 CA2633661 |
189 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA354654110 rs1467011750 |
193 | H>R | No |
ClinGen TOPMed |
|
|
rs1233659440 CA354654130 |
196 | Q>R | No |
ClinGen gnomAD |
|
| TCGA novel | 200 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2633664 rs766816380 |
202 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs900756685 CA354654171 |
202 | S>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs900756685 CA83833970 |
202 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
CA354654179 rs1202578117 |
203 | S>L | No |
ClinGen gnomAD |
|
|
rs1560056772 CA354654187 |
205 | N>D | No |
ClinGen Ensembl |
|
|
CA2633667 rs781754743 |
206 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2633668 rs367665146 |
207 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1490311872 CA354654208 |
208 | E>K | No |
ClinGen gnomAD |
|
|
CA2633670 rs778419590 |
212 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 215 | D>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA354654271 rs1408693472 |
216 | V>A | No |
ClinGen gnomAD |
|
|
rs555454382 CA2633673 |
218 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA354654284 rs1370596861 |
218 | D>Y | No |
ClinGen gnomAD |
|
|
rs1310640060 CA354654327 |
224 | E>A | No |
ClinGen gnomAD |
|
|
CA354654349 rs1370808990 |
227 | P>A | No |
ClinGen TOPMed |
|
| TCGA novel | 227 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2633674 rs746880173 |
229 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA354654365 rs1356966841 |
229 | W>* | No |
ClinGen gnomAD |
|
|
CA354654368 rs746880173 |
229 | W>C | No |
ClinGen ExAC gnomAD |
|
|
rs768475356 CA2633675 |
230 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA354654391 rs1427090006 |
232 | C>F | No |
ClinGen gnomAD |
|
|
CA354654420 rs1275210532 |
236 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA354654419 rs1275210532 |
236 | N>T | No |
ClinGen TOPMed gnomAD |
|
|
CA354654425 rs1342886093 |
237 | G>C | No |
ClinGen gnomAD |
|
| TCGA novel | 237 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs371880420 CA2633677 |
238 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA354654441 rs1486759730 |
239 | V>A | No |
ClinGen gnomAD |
|
|
CA2633678 rs544403184 |
239 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2633680 rs763241416 |
242 | V>E | No |
ClinGen ExAC gnomAD |
|
|
CA354654456 rs1398704013 |
242 | V>L | No |
ClinGen TOPMed |
|
|
CA2633682 rs774572688 |
249 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs759875342 CA354654510 |
250 | M>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759875342 CA2633683 |
250 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1433627253 CA354654507 |
250 | M>V | No |
ClinGen gnomAD |
|
|
rs767790090 CA2633684 |
254 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs376643488 CA2633685 |
257 | S>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
RCV000956139 rs72978714 CA2633687 |
258 | G>R | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA2633688 rs749985486 |
258 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA354654586 rs1301313850 |
261 | P>T | No |
ClinGen gnomAD |
|
|
CA354654598 rs1576995007 |
262 | S>* | No |
ClinGen Ensembl |
|
|
rs1203348089 CA354654605 |
263 | P>S | No |
ClinGen TOPMed |
|
|
CA354654612 rs1201212769 |
264 | P>T | No |
ClinGen gnomAD |
|
|
CA83834057 rs762707873 |
265 | Q>P | No |
ClinGen Ensembl |
|
|
rs202237179 CA2633693 |
266 | C>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2633692 rs754752026 |
266 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1011409073 CA83834082 |
267 | D>G | No |
ClinGen TOPMed |
|
|
rs1454199299 CA354654647 |
268 | Y>F | No |
ClinGen gnomAD |
|
|
rs748086365 CA2633694 |
268 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA354654651 rs1347802436 |
269 | I>L | No |
ClinGen TOPMed |
|
|
CA354654654 rs1304743375 |
269 | I>N | No |
ClinGen TOPMed |
|
| TCGA novel | 270 | R>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs945303800 CA83834087 |
271 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs868431322 CA83834089 |
273 | L>F | No |
ClinGen Ensembl |
|
|
rs1396318188 CA354654698 |
274 | T>I | No |
ClinGen gnomAD |
|
|
rs1478153718 CA354654728 |
277 | F>L | No |
ClinGen gnomAD |
|
| TCGA novel | 278 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1157570636 CA354654743 |
279 | G>V | No |
ClinGen gnomAD |
|
|
COSM728219 rs773408330 CA2633696 |
280 | N>S | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
| TCGA novel | 282 | W>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1485629605 CA354654794 |
285 | G>D | No |
ClinGen Ensembl |
|
|
CA354654804 rs1393707525 |
286 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA354654803 rs1393707525 |
286 | K>T | No |
ClinGen TOPMed gnomAD |
|
|
CA354654820 rs1279246171 |
288 | T>S | No |
ClinGen TOPMed |
|
|
CA2633697 rs749443818 |
289 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs771135953 CA2633698 |
294 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2633699 rs774578403 |
295 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs759702549 CA2633700 |
295 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA354654895 rs1348762636 |
299 | R>G | No |
ClinGen gnomAD |
|
|
CA2633703 rs761148513 |
301 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1374799102 CA354654960 |
308 | R>C | No |
ClinGen TOPMed |
|
|
rs749932433 CA2633705 |
308 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA354654975 rs1416000800 |
310 | S>T | No |
ClinGen TOPMed |
|
|
rs757837731 CA2633708 |
312 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
rs866237155 CA83834161 |
313 | S>L | No |
ClinGen TOPMed gnomAD |
|
|
CA354655012 rs1315256919 |
314 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs373406327 CA2633736 |
314 | P>S | No |
ClinGen ESP ExAC gnomAD |
|
|
CA2633737 rs769014288 |
315 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1031570899 CA83835133 |
316 | D>V | No |
ClinGen Ensembl |
|
|
CA83835140 rs111705120 |
318 | S>P | No |
ClinGen Ensembl |
|
|
rs148197442 CA2633739 |
319 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs148197442 CA2633740 |
319 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs751078214 CA2633741 |
320 | S>* | No |
ClinGen ExAC gnomAD |
|
|
rs759209647 CA2633742 |
323 | A>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 323 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA354655070 rs1484480725 |
324 | Q>P | No |
ClinGen TOPMed gnomAD |
|
|
rs752399185 CA2633744 |
325 | G>E | No |
ClinGen ExAC gnomAD |
|
|
CA2633743 rs767138188 |
325 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1268322419 CA354655090 |
327 | N>T | No |
ClinGen TOPMed |
|
|
rs1190879023 CA354655099 |
328 | K>R | No |
ClinGen gnomAD |
|
|
CA354655139 rs1169297189 |
333 | Q>H | No |
ClinGen gnomAD |
|
|
rs990154157 CA83835170 |
333 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA83835173 rs1002347939 |
334 | L>Q | No |
ClinGen TOPMed |
|
| TCGA novel | 335 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2633747 rs753789797 |
336 | E>D | No |
ClinGen ExAC |
|
|
CA2633748 rs757304667 |
337 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs1265428239 CA354655174 |
339 | Y>C | No |
ClinGen TOPMed |
|
|
rs1382177282 CA354655189 |
341 | I>T | No |
ClinGen gnomAD |
|
|
rs745994136 CA2633750 |
341 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs780075424 CA2633752 |
343 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA354655207 rs1228584580 |
344 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA2633753 rs747247914 |
344 | R>H | No |
ClinGen ExAC |
|
|
rs768967319 CA2633754 |
349 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1211755026 CA354655259 |
351 | E>A | No |
ClinGen gnomAD |
|
|
rs1342717224 CA354655280 |
354 | E>G | No |
ClinGen TOPMed |
|
|
CA2633755 rs777018926 |
355 | H>R | No |
ClinGen ExAC |
|
|
CA83835205 rs931985641 |
357 | K>I | No |
ClinGen Ensembl |
|
|
CA2633756 rs762285377 |
358 | K>M | No |
ClinGen ExAC |
|
|
rs1256394288 CA354655321 |
360 | P>S | No |
ClinGen gnomAD |
|
|
rs1203128364 CA354655325 |
361 | I>V | No |
ClinGen gnomAD |
|
|
rs1249895490 CA354655349 |
364 | S>N | No |
ClinGen gnomAD |
|
|
CA354655354 rs1449991354 |
365 | E>K | No |
ClinGen gnomAD |
|
|
rs1366244170 CA354655368 |
366 | Q>H | No |
ClinGen TOPMed |
|
|
CA2633758 rs773904004 |
369 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA354655394 rs1322776386 |
370 | L>S | No |
ClinGen TOPMed |
|
|
rs150271859 CA2633762 |
371 | Y>* | No |
ClinGen ESP ExAC gnomAD |
|
|
rs752345876 CA2633761 |
371 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs527825012 CA2633760 |
371 | Y>H | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 372 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA354655411 rs1287477128 |
373 | V>I | No |
ClinGen gnomAD |
|
|
CA354655416 rs1436382740 |
374 | K>E | No |
ClinGen gnomAD |
|
|
rs1324675069 CA354655420 |
374 | K>R | No |
ClinGen gnomAD |
|
|
rs1056520181 CA83835247 |
378 | S>W | No |
ClinGen TOPMed |
No associated diseases with P16333
8 regional properties for P16333
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH2 domain | 280 - 376 | IPR000980 |
| domain | SH3 domain | 2 - 61 | IPR001452-1 |
| domain | SH3 domain | 106 - 165 | IPR001452-2 |
| domain | SH3 domain | 190 - 252 | IPR001452-3 |
| domain | Nck1, SH3 domain 1 | 3 - 61 | IPR035562 |
| domain | Nck1, SH3 domain 2 | 108 - 162 | IPR035564 |
| domain | Nck1, SH3 domain 3 | 193 - 249 | IPR035565 |
| domain | Nck1, SH2 domain | 280 - 376 | IPR035882 |
Functions
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein phosphatase type 1 complex | A protein complex that possesses magnesium-dependent protein serine/threonine phosphatase (AMD phosphatase) activity, and consists of a catalytic subunit and one or more regulatory subunits that dictates the phosphatase's substrate specificity, function, and activity. |
| ribosome | An intracellular organelle, about 200 A in diameter, consisting of RNA and protein. It is the site of protein biosynthesis resulting from translation of messenger RNA (mRNA). It consists of two subunits, one large and one small, each containing only protein and RNA. Both the ribosome and its subunits are characterized by their sedimentation coefficients, expressed in Svedberg units (symbol: S). Hence, the prokaryotic ribosome (70S) comprises a large (50S) subunit and a small (30S) subunit, while the eukaryotic ribosome (80S) comprises a large (60S) subunit and a small (40S) subunit. Two sites on the ribosomal large subunit are involved in translation, namely the aminoacyl site (A site) and peptidyl site (P site). Ribosomes from prokaryotes, eukaryotes, mitochondria, and chloroplasts have characteristically distinct ribosomal proteins. |
| vesicle membrane | The lipid bilayer surrounding any membrane-bounded vesicle in the cell. |
12 GO annotations of molecular function
| Name | Definition |
|---|---|
| cadherin binding | Binding to cadherin, a type I membrane protein involved in cell adhesion. |
| cytoskeletal anchor activity | The binding activity of a protein that brings together a cytoskeletal protein (either a microtubule or actin filament, spindle pole body, or protein directly bound to them) and one or more other molecules, permitting them to function in a coordinated way. |
| ephrin receptor binding | Binding to an ephrin receptor. |
| eukaryotic initiation factor eIF2 binding | Binding to eukaryotic initiation factor eIF2, a protein complex involved in the initiation of ribosome-mediated translation. |
| molecular condensate scaffold activity | Binding and bringing together two or more macromolecules in contact, permitting those molecules to organize as a molecular condensate. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase inhibitor activity | Binds to and stops, prevents or reduces the activity of a protein kinase, an enzyme which phosphorylates a protein. |
| protein-macromolecule adaptor activity | The binding activity of a protein that brings together two or more macromolecules in contact, permitting those molecules to function in a coordinated way. The adaptor can bring together two proteins, or a protein and another macromolecule such as a lipid or a nucleic acid. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| signaling adaptor activity | The binding activity of a molecule that brings together two or more molecules in a signaling pathway, permitting those molecules to function in a coordinated way. Adaptor molecules themselves do not have catalytic activity. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| signaling receptor complex adaptor activity | The binding activity of a molecule that provides a physical support for the assembly of a multiprotein receptor signaling complex. |
25 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| lamellipodium assembly | Formation of a lamellipodium, a thin sheetlike extension of the surface of a migrating cell. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | Any process that stops, prevents or reduces the frequency, rate or extent of endoplasmic reticulum stress-induced eiF2alpha phosphorylation. |
| negative regulation of insulin receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of insulin receptor signaling. |
| negative regulation of peptidyl-serine phosphorylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the phosphorylation of peptidyl-serine. |
| negative regulation of PERK-mediated unfolded protein response | Any process that stops, prevents or reduces the frequency, rate or extent of the PERK-mediated unfolded protein response. |
| negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription from an RNA polymerase II promoter as a result of an endoplasmic reticulum stress. |
| peptidyl-serine dephosphorylation | The removal of phosphoric residues from peptidyl-O-phospho-L-serine to form peptidyl-serine. |
| positive regulation of actin filament polymerization | Any process that activates or increases the frequency, rate or extent of actin polymerization. |
| positive regulation of cap-dependent translational initiation | Any process that activates or increases the frequency, rate or extent of cap-dependent translational initiation. |
| positive regulation of cap-independent translational initiation | Any process that activates or increases the frequency, rate or extent of cap-independent translational initiation. |
| positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | Any process that activates or increases the frequency, rate or extent of an endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| positive regulation of T cell proliferation | Any process that activates or increases the rate or extent of T cell proliferation. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of translation in response to endoplasmic reticulum stress | Any process that activates, or increases the frequency, rate or extent of translation as a result of endoplasmic reticulum stress. |
| regulation of cell migration | Any process that modulates the frequency, rate or extent of cell migration. |
| response to other organism | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism. |
| signal complex assembly | The aggregation, arrangement and bonding together of a set of components to form a complex capable of relaying a signal within a cell. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| substrate-dependent cell migration, cell extension | The formation of a cell surface protrusion, such as a lamellipodium or filopodium, at the leading edge of a migrating cell. |
| T cell activation | The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O43639 | NCK2 | Cytoplasmic protein NCK2 | Homo sapiens (Human) | EV |
| P46108 | CRK | Adapter molecule crk | Homo sapiens (Human) | EV |
| P46109 | CRKL | Crk-like protein | Homo sapiens (Human) | SS |
| O55033 | Nck2 | Cytoplasmic protein NCK2 | Mus musculus (Mouse) | SS |
| Q99M51 | Nck1 | Cytoplasmic protein NCK1 | Mus musculus (Mouse) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAEEVVVVAK | FDYVAQQEQE | LDIKKNERLW | LLDDSKSWWR | VRNSMNKTGF | VPSNYVERKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SARKASIVKN | LKDTLGIGKV | KRKPSVPDSA | SPADDSFVDP | GERLYDLNMP | AYVKFNYMAE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| REDELSLIKG | TKVIVMEKCS | DGWWRGSYNG | QVGWFPSNYV | TEEGDSPLGD | HVGSLSEKLA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AVVNNLNTGQ | VLHVVQALYP | FSSSNDEELN | FEKGDVMDVI | EKPENDPEWW | KCRKINGMVG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LVPKNYVTVM | QNNPLTSGLE | PSPPQCDYIR | PSLTGKFAGN | PWYYGKVTRH | QAEMALNERG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HEGDFLIRDS | ESSPNDFSVS | LKAQGKNKHF | KVQLKETVYC | IGQRKFSTME | ELVEHYKKAP |
| 370 | |||||
| IFTSEQGEKL | YLVKHLS |