P16298
Gene name |
PPP3CB (CALNA2, CALNB, CNA2) |
Protein name |
Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform |
Names |
CAM-PRP catalytic subunit, Calmodulin-dependent calcineurin A subunit beta isoform, CNA beta |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5532 |
EC number |
3.1.3.16: Phosphoric monoester hydrolases |
Protein Class |
SERINE/THREONINE-PROTEIN PHOSPHATASE 2B CATALYTIC SUBUNIT 1-RELATED (PTHR45673) |
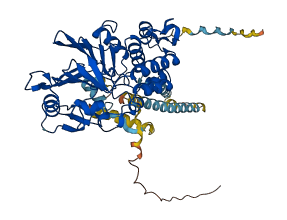
Descriptions
Calcium-dependent, calmodulin-stimulated protein phosphatase (PPP3CB) plays a role in the transduction of intracellular Ca2+-dependent signals in various cellular processes such as cardiac hypertrophy and T cell activation. This protein contains a novel autoinhibitory segment (AIS) in addition to the well-known autoinhibitory domain (AID). AIS and AID cooperatively inhibit enzyme activity.
Autoinhibitory domains (AIDs)
Target domain |
93-293 (Calcineurin-like phosphoesterase domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Target domain |
93-293 (Calcineurin-like phosphoesterase domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
Autoinhibited structure
Activated structure
4 structures for P16298
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4OR9 | X-ray | 223 A | A | 1-524 | PDB |
| 4ORA | X-ray | 275 A | A | 1-524 | PDB |
| 4ORC | X-ray | 270 A | A | 1-524 | PDB |
| AF-P16298-F1 | Predicted | AlphaFoldDB |
191 variants for P16298
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA594370719 rs1433272269 |
1 | M>? | No |
ClinGen gnomAD |
|
|
CA377212845 rs1160414617 |
2 | A>D | No |
ClinGen gnomAD |
|
|
CA377212841 rs1160414617 |
2 | A>V | No |
ClinGen gnomAD |
|
|
rs1413995828 CA377212820 |
3 | A>G | No |
ClinGen gnomAD |
|
|
rs1423121167 CA377212775 |
6 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1423121167 CA377212777 |
6 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1454870048 CA377212770 |
7 | A>T | No |
ClinGen gnomAD |
|
|
rs1186461795 CA377212735 |
8 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA594370709 rs1466035701 |
8 | R>RCV* | No |
ClinGen gnomAD |
|
|
rs1350196304 CA377212667 |
11 | P>L | No |
ClinGen gnomAD |
|
|
CA377212672 rs1242667807 |
11 | P>S | No |
ClinGen gnomAD |
|
|
rs1440236352 CA377212662 |
12 | P>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1440236352 CA377212652 |
12 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA377212664 rs1209277974 |
12 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs763806354 CA377212615 |
13 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs763806354 CA5555745 |
13 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
rs763806354 CA377212617 |
13 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs113693937 CA377212601 |
14 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs113693937 CA209646262 |
14 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1364461760 CA377212573 |
15 | P>L | No |
ClinGen gnomAD |
|
|
rs760374092 CA5555744 |
16 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA377212567 rs1447907900 |
16 | P>S | No |
ClinGen gnomAD |
|
|
CA377212545 rs1380769173 |
17 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA377212543 rs1403054215 |
17 | P>L | No |
ClinGen TOPMed |
|
|
CA377212544 rs1380769173 |
17 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs773877963 CA5555740 |
20 | P>L | No |
ClinGen ExAC |
|
|
rs1435361561 CA377212475 |
20 | P>S | No |
ClinGen gnomAD |
|
|
CA5555738 rs771417243 |
21 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs763382464 CA5555737 |
22 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs955115258 CA209646152 |
23 | A>G | No |
ClinGen TOPMed |
|
|
CA377212436 rs773558588 |
23 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5555736 rs773558588 |
23 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769033884 CA5555732 |
29 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA377209008 rs1181356023 |
30 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1240894692 CA377208996 |
31 | P>S | No |
ClinGen TOPMed |
|
|
CA377208983 rs1293118814 |
32 | F>L | No |
ClinGen gnomAD |
|
|
rs780182113 CA5555707 |
32 | F>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1026496526 CA209633233 |
34 | P>L | No |
ClinGen Ensembl |
|
|
CA5555706 rs772103811 |
34 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749229440 CA5555705 |
37 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs149808527 CA377208938 COSM1349011 |
37 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs149808527 CA5555704 |
37 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1207029573 CA377208927 |
38 | L>F | No |
ClinGen gnomAD |
|
|
rs1415171094 CA377208903 |
40 | S>F | No |
ClinGen TOPMed |
|
|
CA5555701 rs780975567 |
43 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA5555698 rs573948169 |
46 | L>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs138475102 CA5555699 |
46 | L>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA5555697 rs762418445 |
47 | D>V | No |
ClinGen ExAC gnomAD |
|
|
COSM23147 rs1333688207 CA377208748 |
50 | P>L | skin [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA5555696 rs749981068 |
53 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs200220236 CA209633162 |
53 | D>H | No |
ClinGen Ensembl |
|
|
CA377208701 rs1259419512 |
54 | V>I | No |
ClinGen gnomAD |
|
|
rs760558486 CA209633147 |
56 | K>R | No |
ClinGen Ensembl |
|
|
rs762423389 CA5555694 |
60 | V>G | No |
ClinGen ExAC gnomAD |
|
|
CA377208563 rs1364238701 |
63 | G>S | No |
ClinGen TOPMed |
|
|
CA209633127 rs866491225 |
64 | R>* | No |
ClinGen Ensembl |
|
|
rs1385272859 CA377208546 |
64 | R>L | No |
ClinGen gnomAD |
|
|
CA377208480 rs1386370575 |
69 | I>M | No |
ClinGen gnomAD |
|
|
CA5555692 rs764624100 |
69 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs371896052 CA5555693 |
69 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA209633080 rs1043627776 |
70 | A>T | No |
ClinGen Ensembl |
|
|
rs1290520387 CA377208468 COSM3415240 |
70 | A>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs772163544 CA5555689 |
75 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377208388 COSM381300 rs1465995395 |
77 | G>V | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA209633052 rs1012003058 |
79 | A>V | No |
ClinGen Ensembl |
|
|
rs745892650 CA5555688 |
82 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA377208329 rs1173573690 |
82 | R>W | No |
ClinGen gnomAD |
|
|
CA5555687 rs774509144 |
86 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs769657167 CA5555686 |
87 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA377208224 rs1194657674 |
87 | M>V | No |
ClinGen gnomAD |
|
|
CA377208197 rs1209021609 |
88 | I>V | No |
ClinGen TOPMed |
|
|
rs1260286948 CA377208163 |
89 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1207382771 CA377208153 |
90 | V>L | No |
ClinGen gnomAD |
|
|
CA377208130 rs1486256060 |
91 | E>A | No |
ClinGen gnomAD |
|
|
CA5555685 rs748078649 |
91 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA209633022 rs368077062 |
94 | I>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs762470155 COSM1349009 CA209632677 |
106 | D>Y | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA5555666 rs748064202 |
108 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs776597343 CA5555665 |
111 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377207667 rs1307064637 |
119 | N>S | No |
ClinGen gnomAD |
|
|
CA209632656 rs753926654 |
120 | T>I | No |
ClinGen Ensembl |
|
|
rs1214845353 CA377207652 |
121 | R>* | No |
ClinGen TOPMed |
|
|
rs199899901 CA5555664 |
121 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA209632646 rs74453476 |
129 | V>L | No |
ClinGen Ensembl |
|
|
rs1186369101 CA377207487 |
136 | I>V | No |
ClinGen gnomAD |
|
|
rs1247371570 CA377206779 |
138 | C>S | No |
ClinGen gnomAD |
|
|
rs1342340591 CA377206728 |
141 | Y>H | No |
ClinGen gnomAD |
|
|
rs1326788338 CA377206657 |
144 | V>L | No |
ClinGen TOPMed |
|
|
CA5555629 rs755417639 |
151 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs1199581042 CA377206378 |
158 | G>D | No |
ClinGen gnomAD |
|
|
CA5555627 rs766547904 |
163 | R>K | No |
ClinGen ExAC gnomAD |
|
|
CA377206147 rs1372152615 |
167 | E>G | No |
ClinGen gnomAD |
|
|
rs1429231333 CA377205987 |
174 | E>D | No |
ClinGen gnomAD |
|
|
rs1213117450 CA377204953 |
176 | K>R | No |
ClinGen gnomAD |
|
|
rs1336058802 CA377204878 |
179 | Y>C | No |
ClinGen TOPMed |
|
|
rs1414271750 CA377204853 |
181 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA377204797 rs1435566336 |
184 | Y>C | No |
ClinGen gnomAD |
|
|
rs962196869 CA209628034 |
185 | E>G | No |
ClinGen Ensembl |
|
|
CA209628022 rs892282104 |
187 | C>S | No |
ClinGen TOPMed |
|
|
CA377204722 rs1420181378 |
188 | M>I | No |
ClinGen gnomAD |
|
|
rs753897852 CA5555604 |
188 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1187107298 COSM920309 CA377204628 |
193 | S>C | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1188282358 CA377204474 |
202 | Q>E | No |
ClinGen gnomAD |
|
|
CA377204431 rs1194484544 |
204 | F>V | No |
ClinGen TOPMed |
|
|
CA377204409 rs1258849253 COSM920308 |
205 | L>I | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1032604343 CA209628018 |
215 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA5555602 rs759556518 |
216 | H>N | No |
ClinGen ExAC gnomAD |
|
|
CA377204122 rs759556518 |
216 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1448901517 CA377204037 |
220 | D>A | No |
ClinGen gnomAD |
|
|
CA5555601 rs774219136 |
220 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA377204001 rs1274385408 |
222 | R>G | No |
ClinGen gnomAD |
|
|
rs1233179084 CA377203990 |
222 | R>K | No |
ClinGen gnomAD |
|
|
rs1564561831 CA377203978 |
223 | R>G | No |
ClinGen Ensembl |
|
|
rs1472878143 CA377203725 |
225 | D>G | No |
ClinGen TOPMed |
|
|
CA377203639 rs1234014189 |
228 | K>R | No |
ClinGen gnomAD |
|
|
CA209627893 rs202139360 |
229 | E>D | No |
ClinGen TOPMed |
|
|
CA377203448 rs1340780399 |
235 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs373930579 CA377203414 |
236 | M>L | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA209627886 rs373930579 |
236 | M>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA209627862 rs755603004 |
250 | N>H | No |
ClinGen Ensembl |
|
|
CA209627857 rs200953530 |
250 | N>S | No |
ClinGen gnomAD |
|
|
rs751602460 CA5555580 |
251 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA209627845 rs956430664 |
255 | E>A | No |
ClinGen TOPMed |
|
|
CA209627841 rs973638905 |
256 | H>R | No |
ClinGen Ensembl |
|
|
CA377202798 rs1361529412 |
260 | N>S | No |
ClinGen gnomAD |
|
|
CA209627840 rs962312824 |
263 | R>Q | No |
ClinGen Ensembl |
|
|
rs1141282 CA209627839 COSM427884 |
266 | S>F | breast [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA209627837 rs1134796 |
267 | Y>F | No |
ClinGen Ensembl |
|
|
CA377202592 rs1422367208 |
271 | Y>F | No |
ClinGen TOPMed |
|
|
CA209627771 rs1023387956 |
272 | P>S | No |
ClinGen TOPMed |
|
|
rs1374511730 CA377202561 |
273 | A>T | No |
ClinGen gnomAD |
|
|
rs777893115 CA5555563 |
274 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA377202385 rs1397264033 |
280 | N>S | No |
ClinGen gnomAD |
|
|
rs1410869274 CA377202113 |
292 | A>S | No |
ClinGen TOPMed |
|
|
CA377201926 rs1484234762 |
302 | K>Q | No |
ClinGen TOPMed |
|
|
CA377201841 rs1254900458 |
309 | P>R | No |
ClinGen TOPMed |
|
|
CA377201797 rs1416014960 |
312 | I>M | No |
ClinGen gnomAD |
|
|
CA5555539 COSM1349006 rs757043989 |
316 | S>L | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC |
|
rs1327709109 CA377201714 |
319 | N>D | No |
ClinGen gnomAD |
|
|
CA377201654 rs1170061261 |
323 | V>I | No |
ClinGen TOPMed |
|
|
rs1391228775 CA377201630 |
325 | N>H | No |
ClinGen gnomAD |
|
|
CA209625845 rs200425456 |
330 | V>I | No |
ClinGen 1000Genomes gnomAD |
|
|
CA377200784 rs200425456 |
330 | V>L | No |
ClinGen 1000Genomes gnomAD |
|
|
CA5555512 rs574786999 |
335 | N>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs766987677 CA5555511 |
336 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs1331815036 CA377200631 |
336 | N>S | No |
ClinGen gnomAD |
|
|
rs751108387 CA5555509 |
341 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1433910572 CA377200018 |
356 | M>I | No |
ClinGen gnomAD |
|
|
rs766744885 CA5555508 |
356 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA377199912 rs1195485046 |
360 | T>M | No |
ClinGen gnomAD |
|
|
CA5555485 rs139659278 |
373 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs762229258 CA5555483 |
375 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA5555482 rs777159200 |
385 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1268928101 CA377196042 |
387 | M>I | No |
ClinGen TOPMed |
|
|
rs565278043 CA5555436 |
397 | S>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA5555435 rs759770575 |
399 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1354218930 CA377194382 |
399 | A>V | No |
ClinGen gnomAD |
|
|
rs751574172 CA5555434 |
401 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA377194299 rs546930537 |
412 | I>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs546930537 CA5555432 |
412 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1359217754 CA377194266 |
416 | A>V | No |
ClinGen gnomAD |
|
|
rs1400216413 CA377194239 |
420 | S>F | No |
ClinGen gnomAD |
|
|
rs1404791686 CA377225483 |
427 | E>A | No |
ClinGen gnomAD |
|
|
rs1236180758 CA377225308 |
445 | V>L | No |
ClinGen TOPMed |
|
|
CA5555413 rs758464886 |
450 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377225226 rs1437538750 |
454 | Q>R | No |
ClinGen gnomAD |
|
|
rs751426214 CA5555385 |
461 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5555386 rs759360490 |
461 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs766002414 CA5555384 |
465 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs61749234 CA5555373 |
468 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1564545041 CA377223185 |
471 | S>F | No |
ClinGen Ensembl |
|
|
rs1305175476 CA377222908 |
483 | K>R | No |
ClinGen TOPMed |
|
|
rs753997606 CA5555370 |
495 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs781230329 CA5555369 |
497 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA209629731 rs956064020 |
500 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1342647320 CA377222343 |
506 | S>Y | No |
ClinGen gnomAD |
|
|
CA5555362 rs756307973 |
510 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs550798579 CA5555361 |
512 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA377222134 rs1167941200 |
513 | T>I | No |
ClinGen gnomAD |
|
|
rs1399380706 CA377222145 |
513 | T>P | No |
ClinGen gnomAD |
|
|
rs372419696 CA5555360 |
514 | E>K | No |
ClinGen ESP ExAC gnomAD |
|
|
rs775907792 CA5555358 |
517 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377222025 rs775907792 |
517 | G>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5555357 rs746120729 |
518 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5555355 rs778959729 |
520 | N>H | No |
ClinGen ExAC gnomAD |
|
|
CA5555354 rs564975960 |
521 | H>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA377221917 rs1257935972 |
522 | T>A | No |
ClinGen gnomAD |
|
|
CA5555353 rs749327107 |
522 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA377221911 rs749327107 |
522 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA209629683 rs1000041539 |
524 | Q>* | No |
ClinGen TOPMed |
|
|
COSM1505706 CA377221875 rs1320067384 |
524 | Q>L | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
No associated diseases with P16298
3 regional properties for P16298
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.3.16 | Phosphoric monoester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR45673 | SERINE/THREONINE-PROTEIN PHOSPHATASE 2B CATALYTIC SUBUNIT 1-RELATED |
| PANTHER Subfamily | PTHR45673:SF7 | SERINE_THREONINE-PROTEIN PHOSPHATASE 2B CATALYTIC SUBUNIT BETA ISOFORM |
| PANTHER Protein Class |
protein phosphatase
protein modifying enzyme |
|
| PANTHER Pathway Category |
T cell activation Calcineurin Wnt signaling pathway Calcineurin B cell activation Calcineurin |
|
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| calcineurin complex | A heterodimeric calcium ion and calmodulin dependent protein phosphatase composed of catalytic and regulatory subunits; the regulatory subunit is very similar in sequence to calmodulin. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| T-tubule | Invagination of the plasma membrane of a muscle cell that extends inward from the cell surface around each myofibril. The ends of T-tubules make contact with the sarcoplasmic reticulum membrane. |
| Z disc | Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein phosphatase activity | Catalysis of the reaction: protein serine/threonine phosphate + H2O = protein serine/threonine + phosphate, dependent on the presence of calcium-bound calmodulin. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| protein dimerization activity | The formation of a protein dimer, a macromolecular structure consists of two noncovalently associated identical or nonidentical subunits. |
| protein phosphatase 2B binding | Binding to a protein phosphatase 2B. |
| protein serine/threonine phosphatase activity | Catalysis of the reaction: protein serine phosphate + H2O = protein serine + phosphate, and protein threonine phosphate + H2O = protein threonine + phosphate. |
25 GO annotations of biological process
| Name | Definition |
|---|---|
| axon extension | Long distance growth of a single axon process involved in cellular development. |
| calcineurin-mediated signaling | Any intracellular signal transduction in which the signal is passed on within the cell by activation of a transcription factor as a consequence of dephosphorylation by Ca(2+)-activated calcineurin. The process begins with calcium-dependent activation of the phosphatase calcineurin. Calcineurin is a calcium- and calmodulin-dependent serine/threonine protein phosphatase with a conserved function in eukaryotic species from yeast to humans. In yeast and fungi, calcineurin regulates stress signaling and cell cycle, and sporulation and virulence in pathogenic fungi. In metazoans, calcineurin is involved in cell commitment, organogenesis and organ development and immune function of T-lymphocytes. By a conserved mechanism, calcineurin phosphatase activates fungal Crz1 and mammalian NFATc by dephosphorylation and translocation of these transcription factors to the nucleus to regulate gene expression. |
| calcineurin-NFAT signaling cascade | Any intracellular signal transduction in which the signal is passed on within the cell by activation of a member of the NFAT protein family as a consequence of NFAT dephosphorylation by Ca(2+)-activated calcineurin. The cascade begins with calcium-dependent activation of the phosphatase calcineurin. Calcineurin dephosphorylates multiple phosphoserine residues on NFAT, resulting in the translocation of NFAT to the nucleus. The cascade ends with regulation of transcription by NFAT. The calcineurin-NFAT cascade lies downstream of many cell surface receptors, including G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) that signal to mobilize calcium ions (Ca2+). |
| calcium-ion regulated exocytosis | The release of intracellular molecules (e.g. hormones, matrix proteins) contained within a membrane-bounded vesicle by fusion of the vesicle with the plasma membrane of a cell, induced by a rise in cytosolic calcium-ion levels. |
| dephosphorylation | The process of removing one or more phosphoric (ester or anhydride) residues from a molecule. |
| heart development | The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. |
| learning | Any process in an organism in which a relatively long-lasting adaptive behavioral change occurs as the result of experience. |
| locomotion involved in locomotory behavior | Self-propelled movement of a cell or organism from one location to another in a behavioral context; the aspect of locomotory behavior having to do with movement. |
| lymphangiogenesis | Lymph vessel formation when new vessels emerge from the proliferation of pre-existing vessels. |
| memory | The activities involved in the mental information processing system that receives (registers), modifies, stores, and retrieves informational stimuli. The main stages involved in the formation and retrieval of memory are encoding (processing of received information by acquisition), storage (building a permanent record of received information as a result of consolidation) and retrieval (calling back the stored information and use it in a suitable way to execute a given task). |
| negative regulation of T cell mediated cytotoxicity | Any process that stops, prevents, or reduces the rate of T cell mediated cytotoxicity. |
| positive regulation of insulin secretion involved in cellular response to glucose stimulus | Any process that increases the frequency, rate or extent of the regulated release of insulin that contributes to the response of a cell to glucose. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of transcription, DNA-templated | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| protein dephosphorylation | The process of removing one or more phosphoric residues from a protein. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of insulin secretion | Any process that modulates the frequency, rate or extent of the regulated release of insulin. |
| regulation of synaptic plasticity | A process that modulates synaptic plasticity, the ability of synapses to change as circumstances require. They may alter function, such as increasing or decreasing their sensitivity, or they may increase or decrease in actual numbers. |
| regulation of synaptic vesicle endocytosis | Any process that modulates the frequency, rate or extent of synaptic vesicle endocytosis. |
| response to cytokine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cytokine stimulus. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| T cell activation | The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
| T cell differentiation | The process in which a precursor cell type acquires characteristics of a more mature T-cell. A T cell is a type of lymphocyte whose definin characteristic is the expression of a T cell receptor complex. |
| T cell homeostasis | The process of regulating the proliferation and elimination of T cells such that the total number of T cells within a whole or part of an organism is stable over time in the absence of an outside stimulus. |
| T cell proliferation | The expansion of a T cell population by cell division. Follows T cell activation. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P23287 | CNA1 | Serine/threonine-protein phosphatase 2B catalytic subunit A1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P48452 | PPP3CA | Protein phosphatase 3 catalytic subunit alpha | Bos taurus (Bovine) | SS |
| Q27889 | Pp2B-14D | Serine/threonine-protein phosphatase 2B catalytic subunit 2 | Drosophila melanogaster (Fruit fly) | SS |
| P48456 | CanA1 | Serine/threonine-protein phosphatase 2B catalytic subunit 1 | Drosophila melanogaster (Fruit fly) | SS |
| Q9VXF1 | CanA-14F | Serine/threonine-protein phosphatase 2B catalytic subunit 3 | Drosophila melanogaster (Fruit fly) | SS |
| P48454 | PPP3CC | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform | Homo sapiens (Human) | SS |
| Q08209 | PPP3CA | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Homo sapiens (Human) | EV |
| P48455 | Ppp3cc | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform | Mus musculus (Mouse) | PR |
| P63328 | Ppp3ca | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Mus musculus (Mouse) | EV |
| P48453 | Ppp3cb | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform | Mus musculus (Mouse) | SS |
| P63329 | Ppp3ca | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Rattus norvegicus (Rat) | SS |
| P20651 | Ppp3cb | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform | Rattus norvegicus (Rat) | SS |
| Q0G819 | tax-6 | Serine/threonine-protein phosphatase 2B catalytic subunit | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAPEPARAA | PPPPPPPPPP | PGADRVVKAV | PFPPTHRLTS | EEVFDLDGIP | RVDVLKNHLV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KEGRVDEEIA | LRIINEGAAI | LRREKTMIEV | EAPITVCGDI | HGQFFDLMKL | FEVGGSPANT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RYLFLGDYVD | RGYFSIECVL | YLWVLKILYP | STLFLLRGNH | ECRHLTEYFT | FKQECKIKYS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ERVYEACMEA | FDSLPLAALL | NQQFLCVHGG | LSPEIHTLDD | IRRLDRFKEP | PAFGPMCDLL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| WSDPSEDFGN | EKSQEHFSHN | TVRGCSYFYN | YPAVCEFLQN | NNLLSIIRAH | EAQDAGYRMY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RKSQTTGFPS | LITIFSAPNY | LDVYNNKAAV | LKYENNVMNI | RQFNCSPHPY | WLPNFMDVFT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| WSLPFVGEKV | TEMLVNVLSI | CSDDELMTEG | EDQFDGSAAA | RKEIIRNKIR | AIGKMARVFS |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VLREESESVL | TLKGLTPTGM | LPSGVLAGGR | QTLQSATVEA | IEAEKAIRGF | SPPHRICSFE |
| 490 | 500 | 510 | 520 | ||
| EAKGLDRINE | RMPPRKDAVQ | QDGFNSLNTA | HATENHGTGN | HTAQ |