P14653
Gene name |
HOXB1 (HOX2I) |
Protein name |
Homeobox protein Hox-B1 |
Names |
Homeobox protein Hox-2I |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:3211 |
EC number |
|
Protein Class |
|
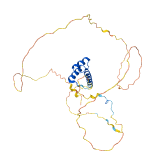
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
207-264 (Homeodomain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for P14653
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1B72 | X-ray | 235 A | A | 170-264 | PDB |
| AF-P14653-F1 | Predicted | AlphaFoldDB |
364 variants for P14653
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000585800 rs1247386618 CA400086893 |
22 | Y>* | Facial paresis, hereditary congenital, 3 Facial paresis, hereditary congenital, 3 (hcfp3) [ClinVar, Ensembl] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000872417 rs150583509 RCV002487917 |
95 | P>T | Facial paresis, hereditary congenital, 3 [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
COSM3937442 rs387907239 VAR_068723 CA129954 RCV000029225 |
207 | R>C | oesophagus Facial paresis, hereditary congenital, 3 Facial paresis, hereditary congenital, 3 (hcfp3) HCFP3; decreased transactivation activity at low DNA concentrations; increased transactivation activity at high DNA concentrations compared to wild-type [Cosmic, ClinVar, Ensembl, UniProt] | Yes |
ClinGen cosmic curated ClinVar UniProt ExAC TOPMed dbSNP gnomAD |
|
RCV000585798 rs1555632121 CA400081746 |
207 | R>H | Facial paresis, hereditary congenital, 3 Facial paresis, hereditary congenital, 3 (hcfp3) [ClinVar, Ensembl] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1567932264 RCV000785946 |
255 | R>Q | Facial paresis, hereditary congenital, 3 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs7226137 VAR_058129 RCV001703117 RCV001692450 RCV001529661 |
265 | E>G | Facial paresis, hereditary congenital, 3 [ClinVar] | Yes |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs1230978425 | 2 | D>A | No |
TOPMed gnomAD |
|
| rs1332752056 | 2 | D>Y | No |
TOPMed gnomAD |
|
| rs763448749 | 3 | Y>C | No |
ExAC gnomAD |
|
| rs1267410884 | 6 | M>I | No | gnomAD | |
| rs765827877 | 6 | M>V | No |
ExAC gnomAD |
|
| rs1598733582 | 7 | N>S | No | gnomAD | |
| rs1598733582 | 7 | N>T | No | gnomAD | |
| rs1226418367 | 7 | N>Y | No |
TOPMed gnomAD |
|
| rs867398927 | 8 | S>F | No |
TOPMed gnomAD |
|
| rs867398927 | 8 | S>Y | No |
TOPMed gnomAD |
|
| rs1279822486 | 10 | L>F | No |
TOPMed gnomAD |
|
| rs760042109 | 10 | L>V | No |
ExAC gnomAD |
|
| rs1421068863 | 13 | P>S | No |
TOPMed gnomAD |
|
| rs570793908 | 14 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201732118 | 14 | L>H | No |
TOPMed gnomAD |
|
| rs201732118 | 14 | L>P | No |
TOPMed gnomAD |
|
| rs570793908 | 14 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2068434736 | 15 | C>R | No | Ensembl | |
| rs1327264822 | 15 | C>Y | No | gnomAD | |
| rs768410617 | 17 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs768410617 | 17 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs550774588 | 17 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs779842484 | 18 | G>* | No |
ExAC TOPMed gnomAD |
|
| rs2068434544 | 18 | G>E | No | TOPMed | |
| rs779842484 | 18 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs769774049 | 19 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs769774049 | 19 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs2144694324 | 19 | P>S | No | Ensembl | |
| rs537215928 | 20 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2068434360 | 21 | A>P | No | gnomAD | |
| rs1251229273 | 21 | A>V | No |
TOPMed gnomAD |
|
| rs12950537 | 22 | Y>H | No |
ExAC gnomAD |
|
| rs200375517 | 24 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200375517 | 24 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
RCV000870526 rs534792734 |
25 | H>missing | No |
ClinVar dbSNP |
|
| rs1218292839 | 25 | H>Q | No | gnomAD | |
|
rs2068433973 COSM3518714 |
25 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs200151818 | 26 | S>R | No | gnomAD | |
| VAR_003817 | 27 | A>AHSA | allele HOXB1*B [UniProt] | No | UniProt |
| rs1305173448 | 27 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1441060785 | 28 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs758818644 | 29 | T>A | No |
ExAC gnomAD |
|
| rs753115367 | 29 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs753115367 | 29 | T>N | No |
ExAC TOPMed gnomAD |
|
|
rs753115367 COSM3362190 |
29 | T>S | kidney [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1347318697 | 30 | S>A | No | gnomAD | |
| rs1393574843 | 31 | F>* | No | gnomAD | |
| rs765599353 | 31 | F>L | No |
ExAC gnomAD |
|
| rs2144694181 | 31 | F>L | No | Ensembl | |
| rs755545003 | 32 | P>R | No |
ExAC gnomAD |
|
| rs1247441581 | 33 | P>L | No | gnomAD | |
| rs1341114446 | 34 | S>R | No | TOPMed | |
| rs1567932867 | 35 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1242617119 | 36 | A>T | No |
TOPMed gnomAD |
|
| rs1401294051 | 38 | A>P | No |
TOPMed gnomAD |
|
| rs1401294051 | 38 | A>T | No |
TOPMed gnomAD |
|
| rs766995509 | 38 | A>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 39 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1282077352 | 40 | D>G | No | gnomAD | |
|
COSM1521291 rs944421558 COSM6146820 |
43 | A>S | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs944421558 | 43 | A>T | No | gnomAD | |
| rs2068432966 | 43 | A>V | No | TOPMed | |
| rs1314713919 | 44 | S>G | No |
TOPMed gnomAD |
|
| rs768986218 | 44 | S>I | No | Ensembl | |
| rs768986218 | 44 | S>N | No | Ensembl | |
| COSM561205 | 45 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4067367 | 46 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201520016 | 47 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs560538130 | 47 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2068432637 | 49 | G>A | No |
TOPMed gnomAD |
|
| rs376245728 | 49 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1315037460 | 50 | G>A | No |
TOPMed gnomAD |
|
| rs1315037460 | 50 | G>E | No |
TOPMed gnomAD |
|
| rs745794647 | 50 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs745794647 | 50 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs771127422 | 51 | G>A | No |
ExAC gnomAD |
|
| COSM4528219 | 51 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1489587559 | 53 | S>F | No |
TOPMed gnomAD |
|
| rs1181184045 | 55 | P>S | No | gnomAD | |
| rs1474113817 | 56 | A>G | No |
TOPMed gnomAD |
|
| rs1474113817 | 56 | A>V | No |
TOPMed gnomAD |
|
|
CA10605593 RCV000352097 rs886043501 |
57 | F>Y | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs1211168663 | 60 | N>D | No | gnomAD | |
| rs1206092299 | 62 | G>D | No | gnomAD | |
| rs779359864 | 62 | G>S | No |
ExAC gnomAD |
|
| rs2068432254 | 63 | Y>D | No | Ensembl | |
| rs1307534184 | 64 | P>L | No | gnomAD | |
|
COSM5681252 rs755351120 |
65 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1353989107 | 68 | P>L | No | gnomAD | |
|
COSM3889818 rs1370259338 |
70 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1370259338 | 70 | S>W | No |
TOPMed gnomAD |
|
|
VAR_055959 rs35254561 |
71 | T>N | No |
UniProt Ensembl dbSNP |
|
| rs1366984237 | 72 | L>P | No | TOPMed | |
| rs1355134865 | 72 | L>V | No |
TOPMed gnomAD |
|
| rs2068431778 | 73 | G>E | No | Ensembl | |
| rs1668267410 | 73 | G>R | No | Ensembl | |
| rs1164029438 | 74 | V>L | No |
TOPMed gnomAD |
|
|
COSM3518712 rs1474149158 |
75 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1419781550 | 76 | F>L | No |
TOPMed gnomAD |
|
| rs763670713 | 77 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs763670713 | 77 | P>S | No |
ExAC TOPMed gnomAD |
|
| COSM74939 | 78 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1343618539 | 78 | S>N | No | gnomAD | |
| rs41306355 | 79 | S>P | No | Ensembl | |
| rs199503961 | 80 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs199503961 | 80 | A>T | No |
ExAC TOPMed gnomAD |
|
|
COSM4067366 rs765050616 |
80 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs776614578 | 81 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs776614578 | 81 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2144693959 | 82 | S>L | No | Ensembl | |
| rs1213632034 | 83 | G>R | No | gnomAD | |
| COSM6081053 | 83 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1275593018 | 84 | Y>C | No | gnomAD | |
|
COSM705968 rs1023657929 |
85 | A>T | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs544516524 | 85 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs530967477 | 86 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1338385831 | 87 | A>T | No | TOPMed | |
| TCGA novel | 88 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779047774 | 88 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2144693906 | 91 | P>L | No | Ensembl | |
| rs1186512882 | 91 | P>S | No | gnomAD | |
| TCGA novel | 92 | S>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1287270276 | 93 | Y>C | No |
TOPMed gnomAD |
|
| rs373253731 | 94 | G>E | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3518711 rs780483178 |
94 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs150583509 | 95 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1285352392 | 97 | Q>R | No | TOPMed | |
| rs777239971 | 98 | Y>* | No |
ExAC TOPMed gnomAD |
|
| COSM6081054 | 100 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757885753 | 101 | L>V | No |
ExAC gnomAD |
|
| rs1191279883 | 102 | G>D | No | gnomAD | |
| rs1191279883 | 102 | G>V | No | gnomAD | |
| rs754648348 | 103 | Q>E | No | Ensembl | |
|
RCV001637730 VAR_055960 rs12939811 |
103 | Q>H | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs764851719 | 103 | Q>R | No |
ExAC gnomAD |
|
| COSM261317 | 105 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs148460064 | 105 | E>D | No |
ESP ExAC gnomAD |
|
| COSM4820393 | 105 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1567932695 | 105 | E>Q | No | Ensembl | |
| rs1484691128 | 106 | G>E | No |
TOPMed gnomAD |
|
| COSM705969 | 107 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1026719724 | 107 | D>G | No | TOPMed | |
| rs773334553 | 108 | G>R | No |
ExAC gnomAD |
|
| rs772165312 | 109 | G>A | No |
ExAC gnomAD |
|
| rs2068430139 | 110 | Y>C | No | TOPMed | |
| rs2068430107 | 111 | F>L | No | TOPMed | |
| rs761868927 | 112 | H>Q | No |
ExAC gnomAD |
|
| COSM3795731 | 113 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2068430049 | 114 | S>* | No |
TOPMed gnomAD |
|
| rs2068430049 | 114 | S>W | No |
TOPMed gnomAD |
|
| rs572890821 | 115 | S>I | No |
1000Genomes ExAC gnomAD |
|
| rs2068429964 | 116 | Y>D | No | gnomAD | |
| rs749602209 | 117 | G>A | No |
ExAC gnomAD |
|
| rs2068429941 | 117 | G>R | No | Ensembl | |
| rs780429744 | 118 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs1008667115 | 118 | A>P | No | Ensembl | |
| rs780429744 | 118 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1454116018 | 120 | L>Q | No |
TOPMed gnomAD |
|
| rs1376755257 | 121 | G>R | No | gnomAD | |
| rs746343075 | 122 | G>A | No |
ExAC gnomAD |
|
| rs890668095 | 123 | L>S | No |
TOPMed gnomAD |
|
| rs2068429623 | 124 | S>F | No | TOPMed | |
| COSM4067365 | 124 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752158237 | 125 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1028968103 | 125 | D>H | No |
TOPMed gnomAD |
|
| rs1028968103 | 125 | D>N | No |
TOPMed gnomAD |
|
| rs752158237 | 125 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs1028968103 | 125 | D>Y | No |
TOPMed gnomAD |
|
| rs778559393 | 126 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1486381791 | 128 | G>E | No |
TOPMed gnomAD |
|
| rs766256495 | 128 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2068429378 | 129 | A>E | No | Ensembl | |
| TCGA novel | 130 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200933753 | 130 | G>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372920233 | 130 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1304170421 | 132 | A>T | No | gnomAD | |
| rs2068429233 | 132 | A>V | No | Ensembl | |
| rs545841960 | 133 | G>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs545841960 | 133 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs545841960 | 133 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2068429112 | 133 | G>V | No | Ensembl | |
| rs774301837 | 134 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs774301837 | 134 | P>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 135 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763137794 | 136 | P>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 136 | P>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1326906734 | 138 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs770139021 | 139 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs530881737 | 140 | Q>R | No | gnomAD | |
| rs902338362 | 141 | H>Q | No | TOPMed | |
| rs2068428744 | 142 | P>H | No |
TOPMed gnomAD |
|
| COSM1383996 | 143 | P>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777134943 | 143 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs771343186 | 144 | Y>H | No |
ExAC gnomAD |
|
| rs368471890 | 146 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs748921455 | 147 | E>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 148 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2144693668 | 149 | T>I | No | Ensembl | |
| rs1486744106 | 150 | A>E | No |
TOPMed gnomAD |
|
| rs750258334 | 150 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs750258334 | 150 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs750258334 | 150 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs757174933 | 151 | S>R | No |
ExAC gnomAD |
|
| rs2068428300 | 153 | A>T | No | TOPMed | |
|
COSM1383995 rs751611344 |
154 | P>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs751611344 | 154 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs2068428268 | 154 | P>S | No | TOPMed | |
| TCGA novel | 155 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1630241 rs1219369123 |
156 | Y>F | liver [Cosmic] | No |
cosmic curated TOPMed |
| rs930945042 | 157 | A>D | No |
TOPMed gnomAD |
|
| rs1354583758 | 157 | A>S | No |
TOPMed gnomAD |
|
| rs930945042 | 157 | A>V | No |
TOPMed gnomAD |
|
| TCGA novel | 159 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 161 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs144625073 RCV000983909 |
162 | E>K | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs144625073 | 162 | E>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs368946771 | 163 | D>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs771417327 | 164 | K>E | No |
ExAC gnomAD |
|
| rs370348373 | 165 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs370348373 | 165 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| COSM3518709 | 165 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1170181865 | 166 | T>I | No |
TOPMed gnomAD |
|
| TCGA novel | 167 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1466541070 | 167 | P>S | No | gnomAD | |
| rs200228944 | 169 | P>A | No |
TOPMed gnomAD |
|
|
rs200228944 COSM3518708 |
169 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| COSM6146822 | 171 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1396834936 | 172 | P>S | No |
TOPMed gnomAD |
|
| rs779796162 | 174 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs779796162 | 174 | T>N | No |
ExAC TOPMed gnomAD |
|
| COSM4067364 | 175 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs138341371 | 176 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs138341371 | 176 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs916641552 | 178 | R>L | No | gnomAD | |
| rs916641552 | 178 | R>P | No | gnomAD | |
|
rs916641552 COSM3421662 |
178 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs948188618 | 178 | R>W | No |
TOPMed gnomAD |
|
| rs2068427333 | 179 | T>N | No | Ensembl | |
| rs1367574149 | 181 | D>A | No | TOPMed | |
| rs780872597 | 181 | D>N | No |
ExAC TOPMed gnomAD |
|
|
rs2068427218 COSM3518707 |
182 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| COSM2148898 | 184 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2068427117 | 186 | K>N | No | Ensembl | |
| rs2068427142 | 186 | K>R | No | TOPMed | |
| TCGA novel | 188 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2068427035 | 190 | P>S | No |
TOPMed gnomAD |
|
| rs2144693525 | 192 | T>I | No | Ensembl | |
| rs369935545 | 193 | A>G | No |
1000Genomes ExAC gnomAD |
|
| rs369935545 | 193 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs771900693 | 195 | V>A | No | ExAC | |
| rs2068423632 | 196 | S>T | No | Ensembl | |
| rs1279175267 | 197 | E>D | No | gnomAD | |
| rs1324089681 | 198 | P>R | No |
TOPMed gnomAD |
|
| rs912799933 | 199 | G>D | No | TOPMed | |
| rs2068423520 | 199 | G>R | No | TOPMed | |
|
rs2068423460 TCGA novel |
200 | L>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs755001724 | 202 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs755001724 | 202 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs376040365 | 203 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
|
rs376040365 COSM980618 |
203 | P>S | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs756390027 | 205 | G>D | No |
ExAC gnomAD |
|
| rs767973578 | 206 | L>P | No |
ExAC gnomAD |
|
| rs1182037537 | 208 | T>N | No | gnomAD | |
| rs2068422997 | 209 | N>S | No | Ensembl | |
| rs1203055487 | 213 | R>G | No | gnomAD | |
| rs2068422900 | 214 | Q>R | No | TOPMed | |
|
COSM3518706 rs776236487 |
221 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs370489961 | 225 | N>S | No |
ESP TOPMed gnomAD |
|
| rs1396751931 | 227 | Y>* | No |
TOPMed gnomAD |
|
| rs200261536 | 229 | S>G | No | gnomAD | |
| rs2068422586 | 229 | S>R | No | Ensembl | |
| rs772889095 | 230 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs879025404 | 230 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1444453992 | 231 | A>S | No | gnomAD | |
| rs2068422487 | 231 | A>V | No | TOPMed | |
| rs759845470 | 232 | R>L | No |
ExAC TOPMed gnomAD |
|
|
rs759845470 COSM3795729 |
232 | R>Q | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1400987484 | 232 | R>W | No |
TOPMed gnomAD |
|
| COSM6081056 | 233 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1459696170 | 234 | V>A | No | gnomAD | |
|
rs768646936 COSM1679893 |
235 | E>D | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs774220542 | 235 | E>V | No |
ExAC gnomAD |
|
| rs2068422237 | 236 | I>V | No | TOPMed | |
| rs1046749207 | 238 | A>T | No |
TOPMed gnomAD |
|
| COSM6146823 | 238 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746058907 | 239 | T>P | No |
ExAC gnomAD |
|
| TCGA novel | 241 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2068422052 | 241 | E>Q | No | TOPMed | |
| rs781620828 | 243 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM4394546 | 244 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2068421976 | 249 | I>V | No | gnomAD | |
| COSM3518703 | 250 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1191811337 | 250 | W>C | No |
TOPMed gnomAD |
|
| rs751975967 | 253 | N>T | No |
ExAC gnomAD |
|
| rs764467860 | 254 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2068421809 | 254 | R>Q | No | Ensembl | |
| COSM6146824 | 256 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758961981 | 260 | K>N | No |
ExAC gnomAD |
|
| rs1252047691 | 260 | K>Q | No | gnomAD | |
| rs1461274057 | 261 | R>C | No |
TOPMed gnomAD |
|
|
COSM472949 rs753190451 |
261 | R>H | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1422685937 COSM3518702 |
262 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
rs375207701 COSM3958482 |
263 | R>* | lung [Cosmic] | No |
cosmic curated 1000Genomes ExAC gnomAD |
| rs772840168 | 263 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs7226137 | 265 | E>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM3518701 | 265 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs7226137 | 265 | E>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1396433565 | 266 | G>R | No | gnomAD | |
| rs373075655 | 267 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs200541812 | 267 | R>W | No | Ensembl | |
| rs1598732645 | 268 | V>A | No | Ensembl | |
| rs768428755 | 269 | P>H | No |
ExAC gnomAD |
|
| rs1393885598 | 270 | P>L | No |
TOPMed gnomAD |
|
| rs1393885598 | 270 | P>Q | No |
TOPMed gnomAD |
|
| rs1393885598 | 270 | P>R | No |
TOPMed gnomAD |
|
| rs749227419 | 270 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs749227419 | 270 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1467570373 | 271 | A>T | No | gnomAD | |
| rs745957228 | 272 | P>L | No |
ExAC gnomAD |
|
| rs769915103 | 272 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs2068420672 | 273 | P>Q | No | TOPMed | |
| rs2068420701 | 273 | P>S | No | Ensembl | |
| rs1222424600 | 274 | G>D | No | Ensembl | |
| rs1219019432 | 276 | P>S | No | gnomAD | |
| rs1354511832 | 277 | K>R | No |
TOPMed gnomAD |
|
| rs1376632156 | 278 | E>G | No | Ensembl | |
| COSM1710411 | 278 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757584432 | 279 | A>E | No | ExAC | |
| rs2068420454 | 280 | A>D | No | gnomAD | |
| rs1567932201 | 281 | G>R | No | Ensembl | |
| rs1567932198 | 282 | D>E | No | Ensembl | |
| rs1203520570 | 282 | D>G | No |
TOPMed gnomAD |
|
| rs2068420398 | 282 | D>N | No | TOPMed | |
|
rs747329214 COSM4067362 |
283 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs778173379 | 284 | S>A | No |
ExAC TOPMed gnomAD |
|
| COSM3795728 | 284 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs778173379 | 284 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs2068420240 | 286 | Q>P | No |
TOPMed gnomAD |
|
| rs758725882 | 287 | S>L | No |
ExAC TOPMed gnomAD |
|
| COSM1383993 | 288 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 289 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1598732577 | 290 | T>P | No | Ensembl | |
| rs145376006 | 291 | S>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1567932171 RCV000711985 |
291 | S>N | No |
ClinVar Ensembl dbSNP |
|
| rs147689555 | 291 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs145376006 | 291 | S>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs761476529 | 292 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1171274369 | 292 | P>S | No |
TOPMed gnomAD |
|
| rs1598732555 | 294 | A>P | No | gnomAD | |
| rs1598732555 | 294 | A>S | No | gnomAD | |
| rs1428323607 | 295 | S>L | No |
TOPMed gnomAD |
|
| rs2068419717 | 296 | P>L | No | TOPMed | |
| rs912529803 | 297 | S>N | No | Ensembl | |
| rs2068419680 | 297 | S>R | No | TOPMed | |
| rs2068419639 | 299 | V>A | No | Ensembl | |
| rs200443029 | 300 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1598732534 | 300 | T>P | No | TOPMed | |
| rs1598732534 | 300 | T>S | No | TOPMed |
1 associated diseases with P14653
[MIM: 614744]: Facial paresis, hereditary congenital, 3 (HCFP3)
A form of facial paresis, a disease characterized by isolated dysfunction of the facial nerve (CN VII). HCFP3 patients are affected by bilateral facial palsy, facial muscle weakness of muscles innervated by CN VII, hearing loss, and strabismus. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of facial paresis, a disease characterized by isolated dysfunction of the facial nerve (CN VII). HCFP3 patients are affected by bilateral facial palsy, facial muscle weakness of muscles innervated by CN VII, hearing loss, and strabismus. . Note=The disease is caused by variants affecting the gene represented in this entry.
4 regional properties for P14653
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Homeodomain | 201 - 265 | IPR001356 |
| conserved_site | Homeobox, conserved site | 236 - 259 | IPR017970 |
| domain | Homeodomain, metazoa | 225 - 236 | IPR020479-1 |
| domain | Homeodomain, metazoa | 240 - 259 | IPR020479-2 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
11 GO annotations of biological process
| Name | Definition |
|---|---|
| anatomical structure formation involved in morphogenesis | The developmental process pertaining to the initial formation of an anatomical structure from unspecified parts. This process begins with the specific processes that contribute to the appearance of the discrete structure and ends when the structural rudiment is recognizable. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome. |
| anterior/posterior pattern specification | The regionalization process in which specific areas of cell differentiation are determined along the anterior-posterior axis. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism. |
| embryonic skeletal system morphogenesis | The process in which the anatomical structures of the skeleton are generated and organized during the embryonic phase. |
| facial nerve structural organization | The process that contributes to the act of creating the structural organization of the facial nerve. This process pertains to the physical shaping of a rudimentary structure. This sensory and motor nerve supplies the muscles of facial expression and the expression and taste at the anterior two-thirds of the tongue. The principal branches are the superficial opthalmic, buccal, palatine and hyomandibular. The main trunk synapses within pterygopalatine ganglion in the parotid gland and this ganglion then gives of nerve branches which supply the lacrimal gland and the mucous secreting glands of the nasal and oral cavities. |
| facial nucleus development | The process whose specific outcome is the progression of the facial nucleus over time, from its formation to the mature structure. |
| pattern specification process | Any developmental process that results in the creation of defined areas or spaces within an organism to which cells respond and eventually are instructed to differentiate. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of DNA-templated transcription | Any process that modulates the frequency, rate or extent of cellular DNA-templated transcription. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| rhombomere 4 development | The process whose specific outcome is the progression of rhombomere 4 over time, from its formation to the mature structure. Rhombomeres are transverse segments of the developing rhombencephalon. Rhombomeres are lineage restricted, express different genes from one another, and adopt different developmental fates. Rhombomeres are numbered in anterior to posterior order. |
| rhombomere 5 development | The process whose specific outcome is the progression of rhombomere 5 over time, from its formation to the mature structure. Rhombomeres are transverse segments of the developing rhombencephalon. Rhombomeres are lineage restricted, express different genes from one another, and adopt different developmental fates. Rhombomeres are numbered in anterior to posterior order. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P23459 | HOXD8 | Homeobox protein Hox-D8 | Gallus gallus (Chicken) | PR |
| A2T6Z0 | HOXB1 | Homeobox protein Hox-B1 | Pan troglodytes (Chimpanzee) | SS |
| P10105 | lab | Homeotic protein labial | Drosophila melanogaster (Fruit fly) | EV |
| P31268 | HOXA7 | Homeobox protein Hox-A7 | Homo sapiens (Human) | SS |
| P49639 | HOXA1 | Homeobox protein Hox-A1 | Homo sapiens (Human) | SS |
| P09022 | Hoxa1 | Homeobox protein Hox-A1 | Mus musculus (Mouse) | SS |
| O08656 | Hoxa1 | Homeobox protein Hox-A1 | Rattus norvegicus (Rat) | SS |
| A2D649 | HOXB1 | Homeobox protein Hox-B1 | Macaca mulatta (Rhesus macaque) | SS |
| Q28IU6 | hoxd1 | Homeobox protein Hox-D1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q8JH55 | hoxb8b | Homeobox protein Hox-B8b | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| F1Q4R9 | meox1 | Homeobox protein MOX-1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDYNRMNSFL | EYPLCNRGPS | AYSAHSAPTS | FPPSSAQAVD | SYASEGRYGG | GLSSPAFQQN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SGYPAQQPPS | TLGVPFPSSA | PSGYAPAACS | PSYGPSQYYP | LGQSEGDGGY | FHPSSYGAQL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GGLSDGYGAG | GAGPGPYPPQ | HPPYGNEQTA | SFAPAYADLL | SEDKETPCPS | EPNTPTARTF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DWMKVKRNPP | KTAKVSEPGL | GSPSGLRTNF | TTRQLTELEK | EFHFNKYLSR | ARRVEIAATL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ELNETQVKIW | FQNRRMKQKK | REREEGRVPP | APPGCPKEAA | GDASDQSTCT | SPEASPSSVT |
| S |