P14644
Gene name |
Pde4c |
Protein name |
cAMP-specific 3',5'-cyclic phosphodiesterase 4C |
Names |
DPDE1 |
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
|
EC number |
3.1.4.53: Phosphoric diester hydrolases |
Protein Class |
|
Descriptions
(Annotation based on sequence homology with Q08499)
PDE4D encodes a cAMP-specific 3',5'-cyclic phosphodiesterase 4D involved in the hydrolysis of cAMP and cGMP, which play important roles in many physiological processes. The autoinhibitory domain of PDE4D is located in the upstream conserved region 2 (UCR2) that inhibits PDE4D activity. Specifically, the α-helical NQVSE[F/Y]ISXTFLD sequence within UCR2 can fold across the catalytic pocket and thus control access to it. In addition, UCR1 can interact with UCR2 to regulate the activity of PDE4.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
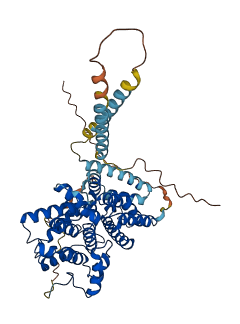
Autoinhibited structure

Activated structure

1 structures for P14644
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P14644-F1 | Predicted | AlphaFoldDB |
No variants for P14644
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P14644 | |||||
1 associated diseases with P14644
[MIM: 617214]: Spermatogenic failure 17 (SPGF17)
An autosomal recessive infertility disorder due to failure of oocyte activation and fertilization by sperm that otherwise exhibits normal morphology. {ECO:0000269|PubMed:26721930}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal recessive infertility disorder due to failure of oocyte activation and fertilization by sperm that otherwise exhibits normal morphology. {ECO:0000269|PubMed:26721930}. Note=The disease is caused by variants affecting the gene represented in this entry.
1 regional properties for P14644
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 5 - 159 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.4.53 | Phosphoric diester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cilium | A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface and of some cytoplasmic parts. Each cilium is largely bounded by an extrusion of the cytoplasmic (plasma) membrane, and contains a regular longitudinal array of microtubules, anchored to a basal body. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| 3',5'-cyclic-AMP phosphodiesterase activity | Catalysis of the reaction: 3',5'-cyclic AMP + H2O = AMP + H+. |
| 3',5'-cyclic-nucleotide phosphodiesterase activity | Catalysis of the reaction: a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate. |
| metal ion binding | Binding to a metal ion. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| cAMP catabolic process | The chemical reactions and pathways resulting in the breakdown of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate). |
| negative regulation of insulin secretion involved in cellular response to glucose stimulus | Any process that decreases the frequency, rate or extent of the regulated release of insulin that contributes to the response of a cell to glucose. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
32 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q28156 | PDE5A | cGMP-specific 3',5'-cyclic phosphodiesterase | Bos taurus (Bovine) | SS |
| P23439 | PDE6B | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Bos taurus (Bovine) | PR |
| P52731 | PDE6C | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' | Gallus gallus (Chicken) | PR |
| H2QL32 | PDE9A | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Pan troglodytes (Chimpanzee) | PR |
| Q9VJ79 | Pde11 | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11 | Drosophila melanogaster (Fruit fly) | SS |
| Q9W4T4 | dnc | 3',5'-cyclic-AMP phosphodiesterase, isoform I | Drosophila melanogaster (Fruit fly) | SS |
| Q9HCR9 | PDE11A | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Homo sapiens (Human) | SS |
| O76083 | PDE9A | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Homo sapiens (Human) | PR |
| Q07343 | PDE4B | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | Homo sapiens (Human) | EV SS |
| Q08499 | PDE4D | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | Homo sapiens (Human) | EV |
| O76074 | PDE5A | cGMP-specific 3',5'-cyclic phosphodiesterase | Homo sapiens (Human) | EV |
| P51160 | PDE6C | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' | Homo sapiens (Human) | PR |
| P27815 | PDE4A | cAMP-specific 3',5'-cyclic phosphodiesterase 4A | Homo sapiens (Human) | EV SS |
| P35913 | PDE6B | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Homo sapiens (Human) | PR |
| Q9Y233 | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Homo sapiens (Human) | PR |
| Q08493 | PDE4C | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Homo sapiens (Human) | EV SS |
| P23440 | Pde6b | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Mus musculus (Mouse) | PR |
| O89084 | Pde4a | 3',5'-cyclic-AMP phosphodiesterase 4A | Mus musculus (Mouse) | SS |
| O70628 | Pde9a | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Mus musculus (Mouse) | PR |
| Q8CG03 | Pde5a | cGMP-specific 3',5'-cyclic phosphodiesterase | Mus musculus (Mouse) | SS |
| Q8CA95 | Pde10a | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Mus musculus (Mouse) | PR |
| P0C1Q2 | Pde11a | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Mus musculus (Mouse) | SS |
| Q01063 | Pde4d | 3',5'-cyclic-AMP phosphodiesterase 4D | Mus musculus (Mouse) | SS |
| Q3UEI1 | Pde4c | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Mus musculus (Mouse) | PR |
| O54735 | Pde5a | cGMP-specific 3',5'-cyclic phosphodiesterase | Rattus norvegicus (Rat) | SS |
| P14270 | Pde4d | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | Rattus norvegicus (Rat) | PR |
| P14646 | Pde4b | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | Rattus norvegicus (Rat) | SS |
| P54748 | Pde4a | 3',5'-cyclic-AMP phosphodiesterase 4A | Rattus norvegicus (Rat) | SS |
| Q8QZV1 | Pde9a | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Rattus norvegicus (Rat) | PR |
| Q8VID6 | Pde11a | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Rattus norvegicus (Rat) | SS |
| Q9QYJ6 | Pde10a | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Rattus norvegicus (Rat) | PR |
| Q22000 | pde-4 | Probable 3',5'-cyclic phosphodiesterase pde-4 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| NSSRTSSAAS | DLHGEDMIVT | PFAQVLASLR | TVRSNVAALA | HGAGSATRQA | LLGTPPQSSQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QAAPAEESGL | QLAQETLEEL | DWCLEQLETL | QTRRSVGEMA | SNKFKRMLNR | ELTHLSETSR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SGNQVSEYIS | QTFLDQQAEV | ELPAPPTEDH | PWPMAQITGL | RKSCHTSLPT | AAIPRFGVQT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DQEEQLAKEL | EDTNKWGLDV | FKVAELSGNR | PLTAVIFRVL | QERDLLKTFQ | IPADTLLRYL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LTLEGHYHSN | VAYHNSIHAA | DVVQSAHVLL | GTPALEAVFT | DLEVLAAIFA | CAIHDVDHPG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VSNQFLINTN | SELALMYNDS | SVLENHHLAV | GFKLLQGENC | DIFQNLSTKQ | KLSLRRMVID |
| 370 | 380 | 390 | 400 | 410 | 420 |
| MVLATDMSKH | MSLLADLKTM | VETKKVTSLG | VLLLDNYSDR | IQVLQSLVHC | ADLSNPAKPL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| PLYRQWTERI | MAEFFQQGDR | ERESGLDISP | MCDKHTASVE | KSQVGFIDYI | AHPLWETWAD |
| 490 | 500 | 510 | 520 | 530 | |
| LVHPDAQELL | DTLEDNREWY | QSRVPCSPPH | AIGPDRFKFE | LTLEETEEEE | EEDERH |