P14633
Gene name |
ecoRIIR |
Protein name |
Type II restriction enzyme EcoRII |
Names |
R.EcoRII, Endonuclease EcoRII, Type-2 restriction enzyme EcoRII |
Species |
Escherichia coli |
KEGG Pathway |
|
EC number |
3.1.21.4: Endodeoxyribonucleases producing 5'-phosphomonoesters |
Protein Class |
|
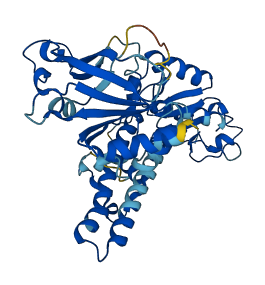
Descriptions
EcoRII is a type IIE restriction endonuclease that interacts with two copies of the DNA recognition sequence 5'-CCWGG, one being the actual target of cleavage, the other serving as the allosteric effector. EcoRII has two functional domains, an N-terminal effector-binding domain and a C-terminal catalytic domain. The catalytic site of EcoRII that is spatially blocked by the N-terminal domain. Activation of the enzyme occurs when an EcoRII dimer interacts with two DNA recognition sites. DNA binding induces domain re-orientation by moving two N-domains away from the catalytic half-sites and moving two C-domains closer to form an active dimer with one substrate DNA duplex. EcoRII cleaves at only one of the two bound recognition sites.
Autoinhibitory domains (AIDs)
Target domain |
229-393 (Catalytic EcoRII domain) |
Relief mechanism |
Ligand binding |
Assay |
Structural analysis |
Target domain |
229-393 (Catalytic EcoRII domain) |
Relief mechanism |
Ligand binding |
Assay |
Structural analysis, Mutagenesis experiment, Deletion assay |
Target domain |
229-393 (Catalytic EcoRII domain) |
Relief mechanism |
Ligand binding |
Assay |
Structural analysis, Mutagenesis experiment, Deletion assay |
Accessory elements
No accessory elements
References
- Zhou XE et al. (2004) "Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold", Journal of molecular biology, 335, 307-19
- Szczepek M et al. (2009) "Molecular analysis of restriction endonuclease EcoRII from Escherichia coli reveals precise regulation of its enzymatic activity by autoinhibition", Molecular microbiology, 72, 1011-21
- Golovenko D et al. (2009) "Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII", Nucleic acids research, 37, 6613-24
Autoinhibited structure
Activated structure
3 structures for P14633
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1NA6 | X-ray | 210 A | A/B | 3-404 | PDB |
| 3HQG | X-ray | 260 A | A | 183-404 | PDB |
| AF-P14633-F1 | Predicted | AlphaFoldDB |
No variants for P14633
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P14633 | |||||
No associated diseases with P14633
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.21.4 | Endodeoxyribonucleases producing 5'-phosphomonoesters |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| type II site-specific deoxyribonuclease activity | Catalysis of the endonucleolytic cleavage of DNA to give specific double-stranded fragments with terminal 5'-phosphates and 3' hydroxyls. Cleavage is dependent on the presence in the DNA of a specific recognition site; cleavage occurs at or very near this recognition site. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| DNA restriction-modification system | A defense process found in many bacteria and archaea that protects the organism from invading foreign DNA by cleaving it with a restriction endonuclease. The organism's own DNA is protected by methylation of a specific nucleotide, which occurs immediately following replication, in the same target site as the restriction enzyme. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MLMSVFHNWL | LEIACENYFV | YIKRLSANDT | GATGGHQVGL | YIPSGIVEKL | FPSINHTREL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NPSVFLTAHV | SSHDCPDSEA | RAIYYNSRHF | GKTRNEKRIT | RWVEAAHFRI | LKITGALTLL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AFKLDEQGGD | CKEVNIWVCA | STDEEDVIET | AIGEVIPGAL | ISGPAGQILG | GLSLQQAPVN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| HKYILPEDWH | LRFPSGSEII | QYAASHYVKN | SLDPDEQLLD | RRRVEYDIFL | LVEELHVLDI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IRKGFGSVDE | FIALANSVSN | RRKSRAGKSL | ELHLEHLFIE | HGLRHFATQA | ITEGNKKPDF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LFPSAGAYHD | TEFPVENLRM | LAVKTTCKDR | WRQILNEADK | IHQVHLFTLQ | EGVSLAQYRE |
| 370 | 380 | 390 | 400 | ||
| MRESGVRLVV | PSSLHKKYPE | AVRAELMTLG | AFIAELTGLY | ADIP |