P14616
Gene name |
INSRR (IRR) |
Protein name |
Insulin receptor-related protein |
Names |
IRR , EC 2.7.10.1 , IR-related receptor [Cleaved into: Insulin receptor-related protein alpha chain; Insulin receptor-related protein beta chain] |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:3645 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
979-1254 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
979-1254 (FnIII-2 domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
1132-1157 (Activation loop from InterPro)
Target domain |
979-1254 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
1132-1157 (Activation loop from InterPro)
Target domain |
979-1254 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Craddock BP et al. (2007) "Autoinhibition of the insulin-like growth factor I receptor by the juxtamembrane region", FEBS letters, 581, 3235-40
- Uchikawa E et al. (2019) "Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex", eLife, 8,
- Nielsen J et al. (2022) "Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling", Journal of molecular biology, 434, 167458
- Chen YS et al. (2021) "Insertion of a synthetic switch into insulin provides metabolite-dependent regulation of hormone-receptor activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Huang X et al. (2009) "Structural insights into the inhibited states of the Mer receptor tyrosine kinase", Journal of structural biology, 165, 88-96
- Hubbard SR (2004) "Juxtamembrane autoinhibition in receptor tyrosine kinases", Nature reviews. Molecular cell biology, 5, 464-71
- Hubbard SR et al. (1994) "Crystal structure of the tyrosine kinase domain of the human insulin receptor", Nature, 372, 746-54
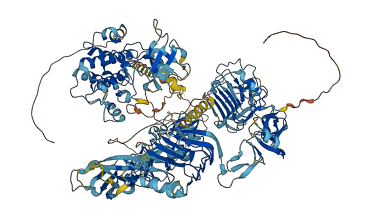
Autoinhibited structure

Activated structure

5 structures for P14616
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 7PL4 | NMR | - | A | 918-948 | PDB |
| 7TYJ | EM | 330 A | A/B | 1-1297 | PDB |
| 7TYK | EM | 350 A | A/B | 1-1297 | PDB |
| 7TYM | EM | 340 A | A/B | 1-1297 | PDB |
| AF-P14616-F1 | Predicted | AlphaFoldDB |
180 variants for P14616
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs56377825 VAR_041437 RCV000986438 RCV002511009 |
246 | C>R | Hereditary insensitivity to pain with anhidrosis [ClinVar] | Yes |
ClinVar UniProt dbSNP |
|
RCV000986437 rs56068937 VAR_041439 |
554 | R>C | Hereditary insensitivity to pain with anhidrosis [ClinVar] | Yes |
ClinVar UniProt dbSNP |
|
rs56252149 RCV000986436 VAR_041440 |
928 | P>L | Hereditary insensitivity to pain with anhidrosis [ClinVar] | Yes |
ClinVar UniProt dbSNP |
|
rs779943666 CA1167726 RCV000708803 RCV001726315 |
1107 | A>S | Familial medullary thyroid carcinoma [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs761392074 RCV000708800 |
1180 | V>missing | Familial medullary thyroid carcinoma [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000986434 rs146201749 |
1254 | F>S | Hereditary insensitivity to pain with anhidrosis [ClinVar] | Yes |
ClinVar dbSNP |
| TCGA novel | 8 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 12 | C>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 13 | L>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs558428940 | 50 | V>M | Variant assessed as Somatic; 0.0006115 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781038209 | 65 | G>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs868120562 | 66 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs143369504 | 69 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs139920561 | 75 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV000917574 rs374390533 |
87 | R>C | No |
ClinVar dbSNP |
|
| rs779432741 | 90 | G>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 91 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756592765 | 96 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs551984020 | 108 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374655019 | 108 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 119 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769822233 | 124 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_041434 rs55757706 |
127 | A>E | No |
UniProt dbSNP |
|
| rs554938935 | 136 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748927042 | 140 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 151 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1442419406 | 156 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs55971900 VAR_041435 |
161 | A>V | No |
UniProt dbSNP |
|
| rs752601283 | 168 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 169 | G>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200875065 | 174 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1347439958 | 190 | P>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 200 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 218 | H>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs140347940 | 232 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 235 | G>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_041436 rs55951840 |
244 | R>H | No |
UniProt dbSNP |
|
| rs752256376 | 244 | R>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369907571 | 250 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 261 | A>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 270 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1399145308 | 271 | S>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751882198 | 273 | R>C | Variant assessed as Somatic; 4.662e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
| VAR_041438 | 278 | E>Q | a lung adenocarcinoma sample; somatic mutation [UniProt] | No | UniProt |
|
RCV000912685 rs150841787 |
279 | R>C | No |
ClinVar dbSNP |
|
| TCGA novel | 282 | S>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754589437 | 289 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1277668841 | 336 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756613475 | 340 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 343 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 346 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770460134 | 359 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 365 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 365 | E>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs184000730 | 367 | Q>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 367 | Q>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 379 | T>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 384 | I>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 410 | G>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 441 | Y>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 451 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 463 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 465 | R>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 496 | A>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 511 | A>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs532604169 | 533 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 539 | G>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 539 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 545 | L>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 553 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1424704699 | 564 | S>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 572 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762538907 | 576 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1199847540 | 577 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs377121387 | 579 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 583 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1275032854 | 583 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 588 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772428423 | 589 | Q>* | Variant assessed as Somatic; 0.000786 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs552398222 | 599 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 635 | L>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 642 | W>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs372272702 | 642 | W>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 646 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 654 | N>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1228589582 | 657 | C>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 659 | R>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759296297 | 669 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751860063 | 688 | C>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 690 | C>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 702 | L>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 711 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 713 | F>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 715 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 716 | F>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 716 | F>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 725 | I>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 728 | W>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777770717 | 730 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755164242 | 739 | R>= | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 747 | A>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 750 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747196691 | 764 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1461335599 | 769 | R>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1272070469 | 786 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 789 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756026000 | 808 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748077768 | 809 | T>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1367479652 | 835 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 844 | G>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 848 | K>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 857 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1462782876 | 867 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767505638 | 869 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748525262 | 875 | G>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 876 | V>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 880 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1004080370 | 882 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1444799871 | 903 | D>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201243410 | 906 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 916 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772259106 | 920 | L>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1255547966 | 923 | L>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1397132963 | 928 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 935 | I>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1368159648 | 945 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 958 | P>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763193926 | 963 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374909614 | 980 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 981 | I>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs190741064 | 983 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745547166 | 989 | S>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1006 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1044 | R>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| VAR_041441 | 1065 | G>E | a glioblastoma multiforme sample; somatic mutation [UniProt] | No | UniProt |
| rs201715497 | 1072 | R>* | Variant assessed as Somatic; 0.0002772 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747555254 | 1072 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs971089803 | 1073 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1074 | L>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779010804 | 1075 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745947130 | 1075 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1086 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1228357495 | 1086 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs145039687 | 1091 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201742278 | 1092 | M>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs982208257 | 1100 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1231811630 | 1101 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs772332648 CA1167723 |
1112 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772332648 CA31081926 |
1112 | V>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA342898948 rs756862919 |
1114 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1167722 rs756862919 |
1114 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1167721 rs753223410 |
1114 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA342898935 rs1403617672 |
1115 | D>Y | No |
ClinGen gnomAD |
|
|
CA342898887 rs1307887055 |
1117 | A>G | No |
ClinGen TOPMed |
|
|
rs777419942 CA1167720 |
1119 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs755716153 CA1167719 |
1119 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA342898849 rs755716153 COSM529505 |
1119 | R>L | lung Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs777419942 CA342898858 |
1119 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1198447148 CA342898814 |
1121 | C>G | No |
ClinGen TOPMed |
|
| TCGA novel | 1134 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780240706 | 1135 | G>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765835617 | 1138 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1039215180 | 1147 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779745536 | 1156 | V>M | Variant assessed as Somatic; 4.62e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs139192917 | 1157 | R>H | Variant assessed as Somatic; 4.62e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1163 | S>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755019469 | 1187 | T>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1190 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1205 | V>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1238 | S>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1241 | H>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1249 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759533268 | 1251 | R>W | Variant assessed as Somatic; 5.056e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs150504965 | 1256 | L>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1263 | P>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1266 | R>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1224959325 | 1268 | A>G | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753459667 | 1268 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780044274 | 1276 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1294 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with P14616
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| insulin receptor complex | A disulfide-bonded, heterotetrameric receptor complex. The alpha chains are entirely extracellular, while each beta chain has one transmembrane domain. The ligand binds to the alpha subunit extracellular domain and the kinase is associated with the beta subunit intracellular domain. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| insulin receptor activity | Combining with insulin receptor ligand and transmitting the signal across the plasma membrane to initiate a change in cell activity. |
| insulin receptor substrate binding | Binding to an insulin receptor substrate (IRS) protein, an adaptor protein that bind to the transphosphorylated insulin and insulin-like growth factor receptors, are themselves phosphorylated and in turn recruit SH2 domain-containing signaling molecules to form a productive signaling complex. |
| phosphatidylinositol 3-kinase binding | Binding to a phosphatidylinositol 3-kinase, any enzyme that catalyzes the addition of a phosphate group to an inositol lipid at the 3' position of the inositol ring. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| cellular response to alkaline pH | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a pH stimulus with pH > 7. pH is a measure of the acidity or basicity of an aqueous solution. |
| male sex determination | The specification of male sex of an individual organism. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
86 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q769I5 | MET | Hepatocyte growth factor receptor | Bos taurus (Bovine) | PR |
| Q05688 | IGF1R | Insulin-like growth factor 1 receptor | Bos taurus (Bovine) | SS |
| A0M8S8 | MET | Hepatocyte growth factor receptor | Felis catus (Cat) (Felis silvestris catus) | PR |
| P09208 | InR | Insulin-like receptor | Drosophila melanogaster (Fruit fly) | SS |
| P08069 | IGF1R | Insulin-like growth factor 1 receptor | Homo sapiens (Human) | EV |
| P08922 | ROS1 | Proto-oncogene tyrosine-protein kinase ROS | Homo sapiens (Human) | SS |
| Q9UM73 | ALK | ALK tyrosine kinase receptor | Homo sapiens (Human) | EV |
| Q06418 | TYRO3 | Tyrosine-protein kinase receptor TYRO3 | Homo sapiens (Human) | SS |
| Q12866 | MERTK | Tyrosine-protein kinase Mer | Homo sapiens (Human) | EV |
| Q01974 | ROR2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Homo sapiens (Human) | EV |
| P07949 | RET | Proto-oncogene tyrosine-protein kinase receptor Ret | Homo sapiens (Human) | EV |
| O15146 | MUSK | Muscle, skeletal receptor tyrosine-protein kinase | Homo sapiens (Human) | EV |
| Q01973 | ROR1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Homo sapiens (Human) | PR |
| Q16832 | DDR2 | Discoidin domain-containing receptor 2 | Homo sapiens (Human) | SS |
| Q08345 | DDR1 | Epithelial discoidin domain-containing receptor 1 | Homo sapiens (Human) | EV |
| Q16620 | NTRK2 | BDNF/NT-3 growth factors receptor | Homo sapiens (Human) | EV |
| P04629 | NTRK1 | High affinity nerve growth factor receptor | Homo sapiens (Human) | EV |
| Q16288 | NTRK3 | NT-3 growth factor receptor | Homo sapiens (Human) | SS |
| P35916 | FLT4 | Vascular endothelial growth factor receptor 3 | Homo sapiens (Human) | SS |
| P17948 | FLT1 | Vascular endothelial growth factor receptor 1 | Homo sapiens (Human) | SS |
| P35968 | KDR | Vascular endothelial growth factor receptor 2 | Homo sapiens (Human) | EV |
| P29317 | EPHA2 | Ephrin type-A receptor 2 | Homo sapiens (Human) | SS |
| P21709 | EPHA1 | Ephrin type-A receptor 1 | Homo sapiens (Human) | SS |
| P54760 | EPHB4 | Ephrin type-B receptor 4 | Homo sapiens (Human) | SS |
| P29376 | LTK | Leukocyte tyrosine kinase receptor | Homo sapiens (Human) | SS |
| P06213 | INSR | Insulin receptor | Homo sapiens (Human) | EV |
| P11362 | FGFR1 | Fibroblast growth factor receptor 1 | Homo sapiens (Human) | EV |
| P21802 | FGFR2 | Fibroblast growth factor receptor 2 | Homo sapiens (Human) | EV |
| P22455 | FGFR4 | Fibroblast growth factor receptor 4 | Homo sapiens (Human) | SS |
| P22607 | FGFR3 | Fibroblast growth factor receptor 3 | Homo sapiens (Human) | EV |
| Q04912 | MST1R | Macrophage-stimulating protein receptor | Homo sapiens (Human) | EV |
| P30530 | AXL | Tyrosine-protein kinase receptor UFO | Homo sapiens (Human) | PR |
| P08581 | MET | Hepatocyte growth factor receptor | Homo sapiens (Human) | EV |
| P04626 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | Homo sapiens (Human) | SS |
| Q15303 | ERBB4 | Receptor tyrosine-protein kinase erbB-4 | Homo sapiens (Human) | SS |
| P21860 | ERBB3 | Receptor tyrosine-protein kinase erbB-3 | Homo sapiens (Human) | SS |
| P00533 | EGFR | Epidermal growth factor receptor | Homo sapiens (Human) | EV |
| P34925 | RYK | Tyrosine-protein kinase RYK | Homo sapiens (Human) | PR |
| Q02763 | TEK | Angiopoietin-1 receptor | Homo sapiens (Human) | SS |
| P35590 | TIE1 | Tyrosine-protein kinase receptor Tie-1 | Homo sapiens (Human) | PR |
| P10721 | KIT | Mast/stem cell growth factor receptor Kit | Homo sapiens (Human) | EV |
| P09619 | PDGFRB | Platelet-derived growth factor receptor beta | Homo sapiens (Human) | EV |
| P07333 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Homo sapiens (Human) | SS |
| P16234 | PDGFRA | Platelet-derived growth factor receptor alpha | Homo sapiens (Human) | EV |
| P36888 | FLT3 | Receptor-type tyrosine-protein kinase FLT3 | Homo sapiens (Human) | EV |
| Q60751 | Igf1r | Insulin-like growth factor 1 receptor | Mus musculus (Mouse) | SS |
| Q62190 | Mst1r | Macrophage-stimulating protein receptor | Mus musculus (Mouse) | SS |
| P97793 | Alk | ALK tyrosine kinase receptor | Mus musculus (Mouse) | SS |
| P15208 | Insr | Insulin receptor | Mus musculus (Mouse) | SS |
| P16056 | Met | Hepatocyte growth factor receptor | Mus musculus (Mouse) | PR |
| Q9WTL4 | Insrr | Insulin receptor-related protein | Mus musculus (Mouse) | SS |
| P97523 | Met | Hepatocyte growth factor receptor | Rattus norvegicus (Rat) | PR |
| P24062 | Igf1r | Insulin-like growth factor 1 receptor | Rattus norvegicus (Rat) | SS |
| P15127 | Insr | Insulin receptor | Rattus norvegicus (Rat) | SS |
| Q64716 | Insrr | Insulin receptor-related protein | Rattus norvegicus (Rat) | SS |
| Q8I7I5 | rol-3 | Protein roller-3 | Caenorhabditis elegans | PR |
| H2KZU7 | svh-2 | Tyrosine-protein kinase receptor svh-2 | Caenorhabditis elegans | SS |
| Q9SXB5 | At1g11303 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11303 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LPZ9 | SD113 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q7X8C5 | WAKL2 | Wall-associated receptor kinase-like 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9S9M2 | WAKL4 | Wall-associated receptor kinase-like 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LMN7 | WAK5 | Wall-associated receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64783 | At1g61370 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64770 | At1g61490 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61490 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SYA0 | At1g61500 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61500 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9C9L5 | WAKL9 | Wall-associated receptor kinase-like 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VYA3 | WAKL10 | Wall-associated receptor kinase-like 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LW83 | CES101 | G-type lectin S-receptor-like serine/threonine-protein kinase CES101 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O81833 | SD11 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M092 | WAKL17 | Wall-associated receptor kinase-like 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64776 | At1g61440 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61440 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SY95 | At1g61550 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61550 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64780 | At1g61400 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61400 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64774 | At1g61460 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61460 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64477 | At2g19130 | G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64782 | SD129 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-29 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64778 | At1g61420 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61420 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SXB4 | At1g11300 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64793 | At1g67520 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64784 | At1g61360 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61360 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q39203 | SD22 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| F1QVU0 | ltk | Tyrosine-protein kinase receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| F8W3R9 | alk | ALK tyrosine kinase receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAVPSLWPWG | ACLPVIFLSL | GFGLDTVEVC | PSLDIRSEVA | ELRQLENCSV | VEGHLQILLM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FTATGEDFRG | LSFPRLTQVT | DYLLLFRVYG | LESLRDLFPN | LAVIRGTRLF | LGYALVIFEM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PHLRDVALPA | LGAVLRGAVR | VEKNQELCHL | STIDWGLLQP | APGANHIVGN | KLGEECADVC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PGVLGAAGEP | CAKTTFSGHT | DYRCWTSSHC | QRVCPCPHGM | ACTARGECCH | TECLGGCSQP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EDPRACVACR | HLYFQGACLW | ACPPGTYQYE | SWRCVTAERC | ASLHSVPGRA | STFGIHQGSC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LAQCPSGFTR | NSSSIFCHKC | EGLCPKECKV | GTKTIDSIQA | AQDLVGCTHV | EGSLILNLRQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GYNLEPQLQH | SLGLVETITG | FLKIKHSFAL | VSLGFFKNLK | LIRGDAMVDG | NYTLYVLDNQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| NLQQLGSWVA | AGLTIPVGKI | YFAFNPRLCL | EHIYRLEEVT | GTRGRQNKAE | INPRTNGDRA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ACQTRTLRFV | SNVTEADRIL | LRWERYEPLE | ARDLLSFIVY | YKESPFQNAT | EHVGPDACGT |
| 550 | 560 | 570 | 580 | 590 | 600 |
| QSWNLLDVEL | PLSRTQEPGV | TLASLKPWTQ | YAVFVRAITL | TTEEDSPHQG | AQSPIVYLRT |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LPAAPTVPQD | VISTSNSSSH | LLVRWKPPTQ | RNGNLTYYLV | LWQRLAEDGD | LYLNDYCHRG |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LRLPTSNNDP | RFDGEDGDPE | AEMESDCCPC | QHPPPGQVLP | PLEAQEASFQ | KKFENFLHNA |
| 730 | 740 | 750 | 760 | 770 | 780 |
| ITIPISPWKV | TSINKSPQRD | SGRHRRAAGP | LRLGGNSSDF | EIQEDKVPRE | RAVLSGLRHF |
| 790 | 800 | 810 | 820 | 830 | 840 |
| TEYRIDIHAC | NHAAHTVGCS | AATFVFARTM | PHREADGIPG | KVAWEASSKN | SVLLRWLEPP |
| 850 | 860 | 870 | 880 | 890 | 900 |
| DPNGLILKYE | IKYRRLGEEA | TVLCVSRLRY | AKFGGVHLAL | LPPGNYSARV | RATSLAGNGS |
| 910 | 920 | 930 | 940 | 950 | 960 |
| WTDSVAFYIL | GPEEEDAGGL | HVLLTATPVG | LTLLIVLAAL | GFFYGKKRNR | TLYASVNPEY |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| FSASDMYVPD | EWEVPREQIS | IIRELGQGSF | GMVYEGLARG | LEAGEESTPV | ALKTVNELAS |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| PRECIEFLKE | ASVMKAFKCH | HVVRLLGVVS | QGQPTLVIME | LMTRGDLKSH | LRSLRPEAEN |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| NPGLPQPALG | EMIQMAGEIA | DGMAYLAANK | FVHRDLAARN | CMVSQDFTVK | IGDFGMTRDV |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| YETDYYRKGG | KGLLPVRWMA | PESLKDGIFT | THSDVWSFGV | VLWEIVTLAE | QPYQGLSNEQ |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| VLKFVMDGGV | LEELEGCPLQ | LQELMSRCWQ | PNPRLRPSFT | HILDSIQEEL | RPSFRLLSFY |
| 1270 | 1280 | 1290 | |||
| YSPECRGARG | SLPTTDAEPD | SSPTPRDCSP | QNGGPGH |