P13535
Gene name |
MYH8 |
Protein name |
Myosin-8 |
Names |
Myosin heavy chain 8 , Myosin heavy chain, skeletal muscle, perinatal , MyHC-perinatal |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4626 |
EC number |
|
Protein Class |
|
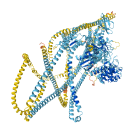
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
82-782 (Myosin head, motor domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P13535
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P13535-F1 | Predicted | AlphaFoldDB |
2192 variants for P13535
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001115657 rs201580344 RCV002556274 RCV001759881 |
20 | R>Q | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000965371 rs146669648 RCV001115655 |
48 | S>F | Hecht syndrome [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000504283 rs375148987 RCV002524229 CA8388455 |
54 | I>T | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001115654 rs2072322757 |
58 | E>Q | Hecht syndrome [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs886052562 CA10648517 RCV000391045 |
68 | G>V | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP |
|
RCV000244704 RCV001120569 rs146732664 CA8388434 RCV000910866 |
75 | R>S | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001120568 rs375714148 RCV002556588 |
85 | P>L | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs144036067 RCV002556587 RCV001120567 RCV003130158 |
93 | M>V | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs149941951 RCV001120566 |
102 | P>A | Hecht syndrome [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs765031203 RCV001120565 |
133 | W>C | Hecht syndrome [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
rs760980150 RCV002502761 RCV000915152 |
160 | I>N | Carney complex - trismus - pseudocamptodactyly syndrome [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA8388313 RCV000502525 rs145863180 RCV000279822 RCV000966459 |
193 | R>H | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs886052561 CA10638830 RCV000374383 |
202 | A>V | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
rs140408926 RCV001560486 RCV001120266 |
226 | A>T | Hecht syndrome [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs753890411 RCV001262909 COSM1479209 |
266 | I>T | Variant assessed as Somatic; MODERATE impact. Hecht syndrome breast [NCI-TCGA, ClinVar, Cosmic] | Yes |
NCI-TCGA Cosmic cosmic curated ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
rs1597404672 RCV001120264 COSM975221 |
284 | R>I | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium Hecht syndrome [NCI-TCGA, Cosmic, ClinVar] | Yes |
NCI-TCGA Cosmic cosmic curated ClinVar Ensembl dbSNP |
|
RCV001116996 rs142606252 RCV000911878 |
318 | Q>H | Hecht syndrome [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs148448600 RCV001116995 |
320 | E>V | Hecht syndrome [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
VAR_050202 RCV000785100 rs34124921 RCV000785101 RCV001116994 |
326 | I>T | Carney complex - trismus - pseudocamptodactyly syndrome Arthrogryposis, distal, type 1A Hecht syndrome [ClinVar] | Yes |
ClinVar UniProt ExAC TOPMed dbSNP gnomAD |
|
rs147828853 RCV001116993 RCV002556480 |
345 | T>S | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001116992 rs759976728 RCV003293886 |
352 | I>L | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
RCV000379745 CA8388164 RCV002521078 rs199851248 |
356 | T>I | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001115567 rs753001993 COSM3402581 |
384 | A>T | Variant assessed as Somatic; MODERATE impact. central_nervous_system Hecht syndrome [NCI-TCGA, Cosmic, ClinVar] | Yes |
NCI-TCGA Cosmic cosmic curated ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV000210629 rs371035884 CA358112 |
422 | Q>* | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC dbSNP |
|
CA8388103 rs747960533 RCV002227152 RCV000322772 |
423 | V>G | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000778486 rs748598841 |
478 | Q>* | Hecht syndrome [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
RCV000778485 rs768299780 |
496 | M>missing | Hecht syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs948334773 RCV001115565 |
528 | E>G | Hecht syndrome [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV001198664 rs149040691 |
547 | T>M | Carney complex - trismus - pseudocamptodactyly syndrome Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
ClinVar 1000Genomes ESP ExAC NCI-TCGA dbSNP gnomAD |
|
RCV000964359 CA8387988 RCV000300494 rs75160168 |
577 | E>D | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000714642 rs760777722 RCV003353005 |
591 | Y>H | Arthrogryposis, distal, type 1A Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
rs1260659117 RCV001120473 |
601 | K>N | Hecht syndrome [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV000927544 rs151091483 RCV001120472 |
605 | N>K | Hecht syndrome [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000117685 rs34693726 RCV001689650 VAR_050203 CA153816 RCV000354138 |
636 | A>V | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs746410520 RCV000778484 |
658 | L>* | Hecht syndrome [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs121434590 VAR_019810 CA123752 RCV000015197 RCV000015198 RCV000438123 |
674 | R>Q | Carney complex - trismus - pseudocamptodactyly syndrome Variant assessed as Somatic; MODERATE impact. Hecht syndrome CACOV and DA7 [ClinVar, NCI-TCGA, UniProt] | Yes |
ClinGen ClinVar UniProt NCI-TCGA TOPMed dbSNP gnomAD |
|
rs150351713 CA8387857 COSM1380755 RCV000369012 |
706 | R>H | large_intestine Hecht syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
CA174419 COSM1179902 RCV000149133 rs193920854 |
780 | R>I | Malignant tumor of prostate prostate [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
rs142137577 RCV002522905 RCV000402897 CA8387690 RCV001356565 |
931 | E>K | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs75477725 RCV000346993 CA8387642 |
996 | S>C | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001120169 rs917329592 |
1051 | L>V | Hecht syndrome [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV000594344 rs751871946 RCV000449642 CA8387544 RCV001199269 |
1107 | L>missing | Carney complex - trismus - pseudocamptodactyly syndrome Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar dbSNP |
|
rs748982401 RCV000320909 CA8387513 |
1126 | E>Q | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000282382 rs138679792 CA8387511 |
1130 | A>S | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs747018022 RCV001115468 |
1157 | G>S | Hecht syndrome [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001115467 rs142073810 CA206127 RCV000192957 |
1178 | R>C | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs372242216 RCV000374507 CA10649454 |
1184 | A>T | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP TOPMed dbSNP |
|
RCV001115465 rs771309686 |
1227 | L>R | Hecht syndrome [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000117689 RCV001115464 rs35962914 CA153824 VAR_050204 RCV002498519 RCV000970024 |
1229 | M>T | Carney complex - trismus - pseudocamptodactyly syndrome Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs199962930 RCV000317571 CA8387472 |
1235 | S>G | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs2072116439 RCV001115463 |
1252 | C>R | Hecht syndrome [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs34953692 RCV001120365 RCV000117690 RCV000973549 CA153826 |
1253 | R>C | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000490328 rs150008607 CA8387419 |
1292 | R>* | Carney complex - trismus - pseudocamptodactyly syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
COSM180550 RCV000260074 CA8387379 rs142232788 |
1335 | A>T | large_intestine Hecht syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA207450 RCV000970023 RCV000193751 RCV002517955 rs140562514 RCV001120363 |
1348 | E>K | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002556580 RCV001120362 rs138607111 |
1383 | I>V | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA10648506 rs886052558 RCV000332157 |
1417 | L>V | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA8387282 RCV000274687 rs201586936 COSM1216248 |
1478 | R>H | large_intestine Hecht syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001120054 COSM1732747 RCV000893692 rs73977155 |
1500 | T>M | pancreas Hecht syndrome [Cosmic, ClinVar] | Yes |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000366956 CA8387217 rs544591533 COSM1216247 |
1563 | R>H | lung large_intestine Hecht syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs61730805 RCV001198663 |
1575 | V>A | Carney complex - trismus - pseudocamptodactyly syndrome [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV000404339 CA8387211 rs181695343 |
1580 | A>T | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA8387206 rs145711576 RCV000363358 |
1595 | R>K | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002541551 rs61730807 RCV000927309 |
1596 | V>I | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001120053 rs139344968 RCV000948447 |
1605 | D>N | Hecht syndrome [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
COSM180547 CA10638823 RCV000306309 rs886052556 |
1637 | R>H | large_intestine Hecht syndrome [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar TOPMed dbSNP gnomAD |
|
CA8387158 rs199865613 RCV000404045 |
1665 | R>Q | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs8069834 CA153830 VAR_030209 RCV000341726 RCV000117692 RCV001647133 |
1692 | W>R | Hecht syndrome [ClinVar] | Yes |
1000Genomes ESP ExAC TOPMed gnomAD ClinGen ClinVar UniProt dbSNP |
|
rs765715898 RCV001118527 |
1732 | K>E | Hecht syndrome [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000989748 RCV000210686 RCV000953038 RCV000194050 CA207949 rs141215006 |
1784 | R>G | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs201598997 CA8387044 RCV000338321 |
1812 | K>R | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001115384 rs148625172 RCV000967437 |
1820 | A>V | Hecht syndrome [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000897671 RCV000249708 rs143876651 CA8387021 RCV000280910 |
1822 | V>I | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000896109 COSM283034 RCV002539443 rs777055080 |
1835 | R>H | large_intestine Inborn genetic diseases [Cosmic, ClinVar] | Yes |
cosmic curated ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs111567318 RCV001115383 RCV001726206 RCV000514232 CA8387011 |
1838 | E>A | Hecht syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000931653 RCV003258761 CA8387001 rs757938047 RCV000373016 |
1847 | E>K | Hecht syndrome Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
COSM2924671 rs376732590 RCV001115382 |
1848 | R>Q | Variant assessed as Somatic; MODERATE impact. Hecht syndrome [NCI-TCGA, ClinVar] | Yes |
NCI-TCGA Cosmic ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV000735361 rs1216879928 |
1915 | I>V | Cognitive impairment [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
| rs2142193591 | 2 | S>N | No | Ensembl | |
| rs1170581501 | 3 | A>G | No |
TOPMed gnomAD |
|
| rs1170581501 | 3 | A>V | No |
TOPMed gnomAD |
|
|
CA398140075 rs1555558639 RCV000522858 |
4 | S>N | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs765461353 | 5 | S>T | No |
ExAC gnomAD |
|
| rs577093992 | 7 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs577093992 | 7 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs373314909 | 9 | M>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs373314909 | 9 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs764647985 | 9 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs775905069 | 10 | A>S | No |
ExAC gnomAD |
|
| rs914097202 | 11 | V>I | No |
TOPMed gnomAD |
|
| rs767784027 | 12 | F>S | No |
ExAC gnomAD |
|
| rs769458253 | 14 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs769458253 | 14 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1313995209 | 15 | A>V | No | gnomAD | |
| rs199744083 | 17 | P>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199744083 | 17 | P>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4063812 rs1331253861 |
18 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs776268681 | 19 | L>I | No |
ExAC gnomAD |
|
|
COSM5893932 rs1449105624 |
20 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs2072324335 | 21 | K>I | No | Ensembl | |
| COSM3513702 | 22 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1230813182 | 23 | E>A | No |
TOPMed gnomAD |
|
| rs746955439 | 26 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs779991062 COSM1520179 COSM6145641 |
26 | R>Q | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
rs746955439 COSM321875 |
26 | R>W | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 27 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1567691394 | 27 | I>T | No | gnomAD | |
| rs758157352 | 28 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs2072323998 | 29 | A>S | No | TOPMed | |
| COSM3889225 | 30 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs377256959 | 30 | Q>E | No | gnomAD | |
| COSM1324194 | 30 | Q>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1597406640 | 31 | N>D | No | Ensembl | |
|
rs753981095 COSM4514864 |
33 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs753981095 | 33 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs757484863 | 33 | P>T | No |
ExAC gnomAD |
|
| rs1217435451 | 34 | F>L | No |
TOPMed gnomAD |
|
| rs1217435451 | 34 | F>V | No |
TOPMed gnomAD |
|
|
COSM6080165 COSM559497 rs1449149762 |
35 | D>E | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs2072323609 | 36 | A>V | No | TOPMed | |
| rs371554777 | 37 | K>E | No |
ESP TOPMed gnomAD |
|
| rs536891720 | 37 | K>T | No |
1000Genomes ExAC gnomAD |
|
| rs939463694 | 38 | T>I | No | Ensembl | |
| rs1300166524 | 39 | S>Y | No |
TOPMed gnomAD |
|
| rs753228332 | 41 | F>I | No |
ExAC gnomAD |
|
| rs2072323413 | 41 | F>L | No | TOPMed | |
| rs1295590545 | 41 | F>S | No | gnomAD | |
| COSM705164 | 42 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1228968956 | 42 | V>M | No |
TOPMed gnomAD |
|
| rs1164448249 | 43 | A>T | No | TOPMed | |
|
rs768031337 COSM975226 |
43 | A>V | endometrium [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs2142193390 | 44 | E>D | No | Ensembl | |
| rs1404447343 | 44 | E>K | No |
TOPMed gnomAD |
|
| rs2072323213 | 45 | P>S | No | Ensembl | |
| rs200490645 | 46 | K>R | No |
1000Genomes ExAC gnomAD |
|
| rs1344093428 | 47 | E>G | No | TOPMed | |
| rs1300867885 | 47 | E>Q | No | Ensembl | |
| rs764857484 | 48 | S>P | No |
ExAC gnomAD |
|
| rs146669648 | 48 | S>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1165311923 | 49 | Y>C | No |
TOPMed gnomAD |
|
| rs1161699264 | 51 | K>T | No |
TOPMed gnomAD |
|
| rs369428507 | 54 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 56 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2142193345 | 56 | S>R | No | Ensembl | |
| rs775327508 | 56 | S>T | No |
ExAC TOPMed gnomAD |
|
| COSM3513699 | 58 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772020544 | 60 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs745704426 | 61 | K>T | No |
ExAC gnomAD |
|
| rs371294844 | 62 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM6145642 | 63 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1321244403 | 63 | T>N | No |
TOPMed gnomAD |
|
|
rs372076449 COSM180559 |
64 | V>I | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs2072322394 | 66 | T>I | No | gnomAD | |
| rs886052562 | 68 | G>D | No | TOPMed | |
| rs2072322375 | 68 | G>S | No | gnomAD | |
| rs749528434 | 69 | G>R | No |
ExAC gnomAD |
|
| rs2142193301 | 69 | G>V | No | Ensembl | |
| rs1210608611 | 71 | T>P | No | TOPMed | |
| TCGA novel | 72 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2142192488 | 72 | L>Q | No | Ensembl | |
| rs373015904 | 72 | L>V | No | gnomAD | |
| rs1019788696 | 73 | T>S | No |
TOPMed gnomAD |
|
| rs2072313203 | 76 | E>K | No |
TOPMed gnomAD |
|
| rs759518186 | 77 | D>V | No |
ExAC gnomAD |
|
| rs1480632533 | 78 | Q>* | No | TOPMed | |
| rs2072313065 | 79 | V>I | No | gnomAD | |
| TCGA novel | 79 | V>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4063811 | 80 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3421274 rs2072312828 |
82 | M>I | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs960013940 | 82 | M>K | No |
TOPMed gnomAD |
|
| rs960013940 | 82 | M>R | No |
TOPMed gnomAD |
|
| rs960013940 | 82 | M>T | No |
TOPMed gnomAD |
|
| rs1369353129 | 83 | N>H | No | gnomAD | |
| rs2072312755 | 83 | N>K | No | TOPMed | |
| rs1207718466 | 84 | P>L | No | gnomAD | |
| TCGA novel | 84 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs375714148 | 85 | P>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1383825542 | 87 | Y>H | No |
TOPMed gnomAD |
|
| COSM1324195 | 88 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4063810 | 88 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1325104252 | 88 | D>N | No | TOPMed | |
| TCGA novel | 89 | K>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs148638810 | 91 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1405863935 | 92 | D>E | No |
TOPMed gnomAD |
|
| rs748236244 | 92 | D>G | No |
ExAC gnomAD |
|
| rs769983204 | 92 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs754946465 | 93 | M>T | No |
ExAC TOPMed gnomAD |
|
|
COSM4827160 rs1230342529 |
95 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| TCGA novel | 96 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072312312 | 96 | M>R | No | TOPMed | |
| rs1483435321 | 98 | H>Q | No |
TOPMed gnomAD |
|
| rs1280253705 | 99 | L>I | No |
TOPMed gnomAD |
|
| rs185811727 | 100 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1044330312 | 100 | H>Y | No | TOPMed | |
| rs1292287529 | 101 | E>K | No | TOPMed | |
| TCGA novel | 102 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs917034420 | 102 | P>R | No | TOPMed | |
| rs149941951 | 102 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs149941951 | 102 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1023778234 | 103 | G>A | No | Ensembl | |
| rs1219272667 | 104 | V>A | No | gnomAD | |
| rs2072311921 | 105 | L>M | No |
TOPMed gnomAD |
|
| rs1324080263 | 106 | Y>* | No | Ensembl | |
| rs1567690832 | 106 | Y>C | No | Ensembl | |
| rs1232271318 | 106 | Y>H | No |
TOPMed gnomAD |
|
| rs2072311776 | 107 | N>D | No | TOPMed | |
| rs139192237 | 107 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072311776 | 107 | N>Y | No | TOPMed | |
| rs1309092986 | 108 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1597406176 | 108 | L>H | No | Ensembl | |
| rs1309092986 | 108 | L>I | No | gnomAD | |
| TCGA novel | 109 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1468156936 | 109 | K>R | No |
TOPMed gnomAD |
|
| rs1164643903 | 110 | E>K | No |
TOPMed gnomAD |
|
| COSM705165 | 110 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3691358 rs940074051 |
111 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
|
COSM1679563 rs908654941 |
111 | R>H | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs908654941 | 111 | R>L | No |
TOPMed gnomAD |
|
| rs2072311513 | 112 | Y>C | No | Ensembl | |
| rs2072311481 | 113 | A>V | No | TOPMed | |
| rs1388811847 | 115 | W>L | No | gnomAD | |
| rs752219854 | 117 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1364071411 | 117 | I>T | No |
TOPMed gnomAD |
|
| rs1597406125 | 119 | T>P | No | Ensembl | |
| rs1188966280 | 120 | Y>H | No | TOPMed | |
| rs768812073 | 120 | Y>S | No |
ExAC gnomAD |
|
| rs200421774 | 122 | G>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200421774 | 122 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs747028184 | 122 | G>S | No |
ExAC gnomAD |
|
| rs1246353008 | 125 | C>F | No | gnomAD | |
| rs2142192211 | 125 | C>G | No | Ensembl | |
| TCGA novel | 125 | C>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1246353008 | 125 | C>S | No | gnomAD | |
| rs1597406115 | 127 | T>P | No | Ensembl | |
| rs752420570 | 128 | V>G | No |
ExAC gnomAD |
|
| rs757502669 | 128 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 129 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754559937 | 129 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs767040391 | 129 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs766363667 | 130 | P>H | No |
ExAC gnomAD |
|
| rs751050296 | 130 | P>T | No |
ExAC gnomAD |
|
| rs544170265 | 131 | Y>* | No |
1000Genomes ExAC gnomAD |
|
| rs377061482 | 131 | Y>C | No |
ESP gnomAD |
|
| rs773068002 | 132 | K>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 133 | W>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV000087202 rs483352720 CA229105 |
133 | W>L | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs761474606 | 135 | P>A | No |
ExAC gnomAD |
|
|
COSM975224 rs768737214 |
135 | P>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs768737214 | 135 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs761474606 | 135 | P>S | No |
ExAC gnomAD |
|
| rs779517713 | 136 | V>G | No |
ExAC gnomAD |
|
| rs376626517 | 136 | V>L | No |
ExAC gnomAD |
|
| rs376626517 | 136 | V>M | No |
ExAC gnomAD |
|
| rs1383572868 | 139 | P>T | No | TOPMed | |
| rs749588346 | 140 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs749588346 | 140 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1597406069 | 141 | V>G | No | Ensembl | |
| rs999815494 | 142 | V>M | No | Ensembl | |
|
RCV000965370 rs138262102 |
144 | A>P | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs138262102 | 144 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs754575329 | 144 | A>V | No |
ExAC gnomAD |
|
| rs1337324222 | 146 | R>G | No | TOPMed | |
| COSM6145643 | 146 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM6080167 | 147 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1456404225 | 148 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs373953153 | 150 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1169419511 | 150 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1169419511 | 150 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1169419511 | 150 | R>P | No |
TOPMed gnomAD |
|
| rs757847581 | 151 | Q>* | No |
ExAC gnomAD |
|
| rs750390854 | 152 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs879016103 | 153 | A>G | No |
TOPMed gnomAD |
|
| rs900231240 | 153 | A>P | No | Ensembl | |
| rs761715060 | 154 | P>L | No |
ExAC gnomAD |
|
| rs1200959028 | 154 | P>S | No | gnomAD | |
| COSM6145644 | 155 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753560889 | 156 | H>P | No |
ExAC TOPMed gnomAD |
|
| rs542453131 | 157 | I>N | No | 1000Genomes | |
| rs895749851 | 157 | I>V | No | TOPMed | |
| rs2072308612 | 158 | F>V | No | TOPMed | |
| rs1343629871 | 159 | S>A | No |
TOPMed gnomAD |
|
| TCGA novel | 159 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 161 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1493787 | 162 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775792268 | 165 | Y>D | No |
ExAC gnomAD |
|
| COSM975222 | 167 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759652599 | 169 | L>F | No |
ExAC gnomAD |
|
| rs2072308164 | 171 | D>N | No | TOPMed | |
|
COSM3513696 rs143063345 |
172 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs571149881 | 172 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1709830 rs571149881 |
172 | R>Q | skin [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1276554003 | 173 | E>D | No | gnomAD | |
| rs2072284625 | 174 | N>T | No | gnomAD | |
| rs1352983479 | 176 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs148334778 | 177 | I>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs148334778 | 177 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 178 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072284513 | 178 | L>P | No | Ensembl | |
| COSM3513695 | 179 | I>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763376361 | 179 | I>T | No |
ExAC gnomAD |
|
| rs143332486 | 180 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs143332486 | 180 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM381200 rs181130030 |
181 | G>R | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| COSM330984 | 182 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1472258546 RCV001531255 |
184 | G>A | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1472258546 | 184 | G>D | No |
TOPMed gnomAD |
|
| TCGA novel | 184 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773625190 | 185 | A>G | No |
ExAC gnomAD |
|
| rs1597405149 | 185 | A>T | No | Ensembl | |
| rs368169632 | 186 | G>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs563940083 | 186 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs368169632 | 186 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072282858 | 187 | K>R | No |
TOPMed gnomAD |
|
| rs768940714 | 189 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs745516575 | 191 | T>A | No |
ExAC gnomAD |
|
| rs1339759887 | 191 | T>N | No | gnomAD | |
| rs1292113272 | 193 | R>C | No | gnomAD | |
| rs145863180 | 193 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 195 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072282634 | 195 | I>V | No | TOPMed | |
| rs1313101303 | 197 | Y>H | No | gnomAD | |
| rs1365595563 | 199 | A>T | No | gnomAD | |
| TCGA novel | 199 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs902439183 | 200 | T>I | No |
TOPMed gnomAD |
|
| rs902439183 | 200 | T>K | No |
TOPMed gnomAD |
|
| rs1351181207 | 201 | I>M | No | gnomAD | |
|
rs766878606 COSM1479211 |
201 | I>T | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs777624162 | 201 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs886052561 | 202 | A>G | No |
TOPMed gnomAD |
|
| COSM705166 | 202 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1404183109 | 204 | T>A | No |
TOPMed gnomAD |
|
| rs1567689658 | 206 | E>D | No | Ensembl | |
| rs781139865 | 207 | K>N | No |
ExAC gnomAD |
|
|
COSM110206 rs144897535 |
209 | K>E | skin [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs963790352 | 210 | D>G | No |
TOPMed gnomAD |
|
| rs751752898 | 210 | D>N | No | ExAC | |
| rs766584008 | 211 | E>* | No |
ExAC gnomAD |
|
|
COSM283037 rs766584008 |
211 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs567013242 | 212 | S>T | No |
TOPMed gnomAD |
|
| rs2072281976 | 213 | G>S | No | TOPMed | |
| rs2072281035 | 217 | G>A | No | TOPMed | |
| rs1192229027 | 217 | G>W | No | gnomAD | |
| rs201516282 | 218 | T>A | No | 1000Genomes | |
| rs1240212475 | 218 | T>I | No | Ensembl | |
| rs201516282 | 218 | T>S | No | 1000Genomes | |
| rs374268509 | 219 | L>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072280860 | 219 | L>P | No | TOPMed | |
| rs374268509 | 219 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs915045580 | 220 | E>K | No |
TOPMed gnomAD |
|
|
COSM1479210 rs915045580 |
220 | E>Q | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| rs2072280799 | 220 | E>V | No | TOPMed | |
| rs2072280750 | 221 | D>N | No | Ensembl | |
| TCGA novel | 221 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM705167 | 222 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1313691005 | 224 | I>L | No |
TOPMed gnomAD |
|
| rs2072280610 | 224 | I>M | No | TOPMed | |
| rs1313691005 | 224 | I>V | No |
TOPMed gnomAD |
|
| rs781228319 | 225 | S>R | No |
ExAC gnomAD |
|
| rs2072280481 | 226 | A>D | No | TOPMed | |
| rs1289668311 | 227 | N>S | No | gnomAD | |
| rs780197977 | 228 | P>S | No |
ExAC gnomAD |
|
| rs758641534 | 229 | L>P | No |
ExAC gnomAD |
|
| rs2072280336 | 230 | L>V | No | Ensembl | |
| rs1356186695 | 233 | F>S | No |
TOPMed gnomAD |
|
| TCGA novel | 234 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072280172 | 235 | N>S | No | TOPMed | |
| COSM3969798 | 236 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs866751698 | 236 | A>T | No | Ensembl | |
| rs2072280151 | 236 | A>V | No | TOPMed | |
| rs754371915 | 239 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs754371915 COSM3370602 |
239 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs760998000 | 241 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs140772576 | 242 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2072279928 | 242 | D>N | No | TOPMed | |
| rs768392636 | 246 | R>C | No |
TOPMed gnomAD |
|
| rs766016765 | 246 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs766016765 | 246 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1425770933 | 248 | G>C | No |
TOPMed gnomAD |
|
| rs2072278507 | 248 | G>D | No | Ensembl | |
| rs929899105 | 252 | R>* | No | Ensembl | |
| rs2072278443 | 252 | R>T | No |
TOPMed gnomAD |
|
| rs1182386288 | 253 | I>T | No |
TOPMed gnomAD |
|
| rs1182760779 | 255 | F>I | No |
TOPMed gnomAD |
|
| rs779063389 | 256 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs757364984 | 258 | T>P | No | ExAC | |
| rs2072278056 | 264 | A>V | No | gnomAD | |
| rs2072278018 | 265 | D>N | No | TOPMed | |
| rs1391523071 | 269 | Y>C | No | gnomAD | |
| rs2072277861 | 269 | Y>H | No | Ensembl | |
|
rs1302815990 COSM4840380 |
270 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs993070610 | 270 | L>P | No | TOPMed | |
| rs781511409 | 272 | E>Q | No |
ExAC gnomAD |
|
| rs755435554 | 274 | S>Y | No |
ExAC gnomAD |
|
| rs753483742 | 278 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs756972191 | 278 | F>Y | No |
ExAC gnomAD |
|
| rs1216199703 | 279 | Q>* | No |
TOPMed gnomAD |
|
| rs1216199703 | 279 | Q>E | No |
TOPMed gnomAD |
|
| rs2072271955 | 279 | Q>H | No | Ensembl | |
| rs1389407671 | 279 | Q>P | No |
TOPMed gnomAD |
|
| rs1597404680 | 280 | L>P | No | TOPMed | |
| rs200214136 | 282 | A>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200214136 | 282 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2072271778 | 283 | E>G | No | gnomAD | |
| rs759382127 | 283 | E>K | No | ExAC | |
|
rs1597404672 COSM3889220 |
284 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs770649624 | 285 | S>N | No |
ExAC gnomAD |
|
| rs774130964 | 285 | S>R | No |
ExAC gnomAD |
|
| rs749530169 | 285 | S>R | No |
ExAC gnomAD |
|
| rs770649624 | 285 | S>T | No |
ExAC gnomAD |
|
| rs1236026136 | 286 | Y>S | No | gnomAD | |
| rs1290966676 | 287 | H>R | No |
TOPMed gnomAD |
|
| rs2072271514 | 288 | I>T | No | TOPMed | |
| rs2072271537 | 288 | I>V | No | Ensembl | |
| rs769949645 | 290 | Y>* | No |
ExAC gnomAD |
|
| rs773262066 | 290 | Y>D | No |
ExAC gnomAD |
|
| rs773262066 | 290 | Y>H | No |
ExAC gnomAD |
|
| rs1319380590 | 290 | Y>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1319380590 COSM5706285 |
290 | Y>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs2072271393 | 291 | Q>L | No |
TOPMed gnomAD |
|
| rs756089815 | 294 | S>F | No | gnomAD | |
| rs1567689246 | 294 | S>P | No | Ensembl | |
| rs923441604 | 295 | N>S | No |
TOPMed gnomAD |
|
| COSM975220 | 296 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs565366982 | 298 | P>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM6145646 | 300 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072271206 | 301 | I>S | No | TOPMed | |
| rs2072271206 | 301 | I>T | No | TOPMed | |
| rs1567689200 | 303 | M>I | No | Ensembl | |
| rs2072270491 | 303 | M>T | No |
TOPMed gnomAD |
|
| rs752133623 | 303 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1470066194 | 304 | L>I | No | gnomAD | |
| rs759476464 | 307 | T>I | No |
ExAC gnomAD |
|
| rs759476464 | 307 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs759476464 | 307 | T>S | No |
ExAC gnomAD |
|
| COSM4935623 | 308 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs150966976 | 310 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs150966976 | 310 | P>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs773523769 | 311 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs150332523 | 312 | D>G | No |
1000Genomes ExAC TOPMed |
|
| rs2072270147 | 312 | D>N | No | TOPMed | |
| rs201581304 | 313 | Y>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776577261 | 313 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1567689174 | 314 | A>S | No |
TOPMed gnomAD |
|
|
rs780665709 COSM1258478 |
316 | V>I | oesophagus [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs780665709 | 316 | V>L | No |
ExAC TOPMed gnomAD |
|
| COSM1679562 | 319 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200409345 | 322 | T>A | No | 1000Genomes | |
| rs1331363277 | 322 | T>I | No | gnomAD | |
| rs1285029288 | 324 | P>A | No | gnomAD | |
| rs1223236011 | 324 | P>L | No | gnomAD | |
| rs777536806 | 325 | S>I | No |
ExAC TOPMed gnomAD |
|
| rs777536806 | 325 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs777536806 | 325 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1381745110 | 328 | D>G | No | gnomAD | |
| rs780695842 | 329 | Q>E | No |
ExAC gnomAD |
|
|
COSM1380758 rs780695842 |
329 | Q>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1468769229 | 329 | Q>P | No |
TOPMed gnomAD |
|
| rs201127112 | 330 | E>G | No | 1000Genomes | |
| rs751532878 | 330 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs766273711 | 333 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs762855645 | 333 | M>T | No |
ExAC gnomAD |
|
| rs750173642 | 335 | T>A | No |
ExAC gnomAD |
|
| rs2072269216 | 336 | D>N | No | TOPMed | |
| rs754523656 | 337 | S>G | No |
ExAC gnomAD |
|
| rs990419920 | 337 | S>N | No | TOPMed | |
| rs1430622800 | 338 | A>V | No |
TOPMed gnomAD |
|
| rs923545568 | 339 | I>T | No |
TOPMed gnomAD |
|
| rs746500718 | 339 | I>V | No |
ExAC gnomAD |
|
| rs780026565 | 340 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1474488948 | 340 | D>N | No | gnomAD | |
| COSM975219 | 342 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200063552 | 344 | F>L | No | 1000Genomes | |
| rs34419805 | 345 | T>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 346 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754152900 | 347 | E>Q | No |
ExAC gnomAD |
|
| COSM1216251 | 348 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764385049 | 349 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs2072267400 | 349 | K>Q | No | TOPMed | |
| rs530350694 | 350 | V>L | No |
1000Genomes ExAC gnomAD |
|
| rs530350694 | 350 | V>M | No |
1000Genomes ExAC gnomAD |
|
| rs1192009068 | 352 | I>N | No |
TOPMed gnomAD |
|
| rs1192009068 | 352 | I>S | No |
TOPMed gnomAD |
|
| rs771234906 | 353 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs774876652 | 353 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1361578332 | 354 | K>N | No |
TOPMed gnomAD |
|
| TCGA novel | 356 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs559977700 | 357 | G>A | No |
1000Genomes ExAC gnomAD |
|
| rs2072266998 | 358 | A>V | No | Ensembl | |
| rs2072266951 | 359 | V>A | No | Ensembl | |
| rs1026216843 | 359 | V>M | No | Ensembl | |
| rs2072266869 | 363 | G>E | No | Ensembl | |
| rs779545922 | 363 | G>R | No |
ExAC gnomAD |
|
| rs1480129894 | 365 | M>T | No | gnomAD | |
| rs958040684 | 366 | K>* | No | TOPMed | |
| rs745822806 | 366 | K>T | No |
ExAC gnomAD |
|
| rs2072266726 | 368 | K>E | No | TOPMed | |
| rs778878392 | 369 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs1480860018 | 372 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs144477514 | 372 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| COSM1324196 | 372 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1480860018 | 372 | R>S | No |
TOPMed gnomAD |
|
| rs1387499907 | 373 | E>K | No | Ensembl | |
| rs1224830966 | 376 | A>S | No | TOPMed | |
| rs1202425013 | 376 | A>V | No | gnomAD | |
| rs372241085 | 379 | D>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072266530 | 379 | D>N | No | TOPMed | |
| rs764474712 | 380 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1363461295 | 385 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1363461295 | 385 | D>N | No | gnomAD | |
| rs2072254972 | 387 | A>G | No | TOPMed | |
| COSM1380757 | 387 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs755066825 | 388 | A>G | No |
ExAC gnomAD |
|
| COSM283036 | 388 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072254849 | 390 | L>F | No | Ensembl | |
| rs2072254849 | 390 | L>I | No | Ensembl | |
| rs1482109939 | 392 | S>N | No | TOPMed | |
| TCGA novel | 393 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072254761 | 394 | N>I | No | gnomAD | |
| COSM975218 | 395 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763456698 | 397 | D>G | No |
ExAC gnomAD |
|
| rs766864218 | 397 | D>N | No |
ExAC gnomAD |
|
| rs1258642830 | 402 | L>F | No | gnomAD | |
|
RCV000449650 rs144321381 CA153812 RCV000117683 |
403 | C>* | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs775053298 | 404 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs888776823 | 407 | V>I | No | gnomAD | |
| rs773912355 | 409 | V>A | No |
ExAC gnomAD |
|
| rs759031617 | 409 | V>I | No | ExAC | |
| rs2072254036 | 410 | G>R | No | TOPMed | |
| COSM3513690 | 410 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749257514 | 411 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs2072253818 | 412 | E>D | No |
TOPMed gnomAD |
|
| rs1290801680 | 412 | E>G | No | TOPMed | |
| rs2072253904 | 412 | E>K | No | Ensembl | |
| rs2072253789 | 413 | Y>H | No | gnomAD | |
| rs777815349 | 416 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs769755030 | 417 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs769755030 | 417 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs781327259 | 418 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2072253455 | 419 | T>I | No | Ensembl | |
| rs1419393171 | 420 | V>M | No | gnomAD | |
| rs751667258 | 421 | Q>H | No |
ExAC gnomAD |
|
| rs747960533 | 423 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs892513199 | 423 | V>L | No |
TOPMed gnomAD |
|
| rs892513199 | 423 | V>M | No |
TOPMed gnomAD |
|
| rs2072252196 | 424 | Y>F | No | TOPMed | |
| rs1371213124 | 425 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1056398923 | 426 | A>S | No | Ensembl | |
| rs150633264 | 426 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs779463903 | 427 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs745934708 | 427 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2142186374 | 428 | G>C | No | Ensembl | |
| rs2072251926 | 429 | A>G | No | Ensembl | |
| COSM705169 | 431 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757761959 | 434 | V>I | No |
ExAC TOPMed gnomAD |
|
|
rs1225449045 COSM1258477 |
436 | E>K | oesophagus [Cosmic] | No |
cosmic curated gnomAD |
| rs1324351336 | 437 | K>N | No | gnomAD | |
| rs764540659 | 438 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1441564754 | 438 | M>V | No | TOPMed | |
|
COSM396440 rs756392331 |
439 | F>L | lung [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
COSM975217 rs141899271 |
440 | L>M | endometrium [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs1329312571 COSM975216 |
440 | L>P | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
|
TCGA novel COSM1493788 rs1225016907 |
441 | W>* | Variant assessed as Somatic; HIGH impact. kidney [NCI-TCGA, Cosmic] | No |
NCI-TCGA cosmic curated Ensembl |
| COSM705170 | 441 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1597403884 | 443 | V>G | No | Ensembl | |
| rs766108256 | 443 | V>L | No |
ExAC gnomAD |
|
| rs762610774 | 444 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs372766209 | 445 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
rs764777507 COSM291879 |
445 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1427980850 | 447 | N>Y | No | gnomAD | |
| rs1049191100 | 448 | Q>H | No |
TOPMed gnomAD |
|
| rs2142186301 | 448 | Q>K | No | Ensembl | |
| rs1173882934 | 449 | Q>K | No | gnomAD | |
| rs776386007 | 450 | L>M | No |
ExAC gnomAD |
|
| rs768534267 | 451 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs2072251181 | 452 | T>N | No | TOPMed | |
| rs1437555604 | 454 | Q>K | No | gnomAD | |
| rs772272975 | 455 | P>A | No |
ExAC gnomAD |
|
|
COSM705171 rs2072251058 |
456 | R>S | Variant assessed as Somatic; MODERATE impact. lung [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs2072251079 | 456 | R>T | No | TOPMed | |
| rs1194836051 | 457 | Q>* | No |
TOPMed gnomAD |
|
| rs1597403857 | 457 | Q>H | No | Ensembl | |
| rs572581326 | 457 | Q>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1262578243 | 459 | F>L | No | gnomAD | |
| rs995889881 | 459 | F>L | No | TOPMed | |
| rs1018737240 | 460 | I>F | No | TOPMed | |
| rs1018737240 | 460 | I>L | No | TOPMed | |
| rs1018737240 | 460 | I>V | No | TOPMed | |
| rs757234101 | 461 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs749830403 | 462 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs749830403 | 462 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs2072250728 TCGA novel |
464 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1404663794 | 465 | I>V | No | TOPMed | |
|
rs554419599 COSM2150563 |
466 | A>V | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
| rs1157145036 | 468 | F>L | No |
TOPMed gnomAD |
|
| rs1338694429 | 468 | F>S | No |
TOPMed gnomAD |
|
|
CA16609484 RCV000593687 rs1060499728 |
470 | I>missing | No |
ClinGen ClinVar dbSNP |
|
| rs756551633 | 472 | D>G | No |
ExAC gnomAD |
|
| rs1453266228 | 472 | D>N | No | gnomAD | |
| rs1417471179 | 477 | E>* | No |
TOPMed gnomAD |
|
| rs781703793 | 478 | Q>R | No |
ExAC gnomAD |
|
| rs2072238680 | 479 | L>M | No | Ensembl | |
| rs891273476 | 480 | C>Y | No |
TOPMed gnomAD |
|
| rs755349321 | 481 | I>V | No |
ExAC gnomAD |
|
| rs184791206 | 482 | N>K | No |
1000Genomes TOPMed |
|
| rs557101124 | 483 | F>C | No | TOPMed | |
| rs2142184819 | 484 | T>N | No | 1000Genomes | |
| rs1242761382 | 485 | N>S | No | gnomAD | |
| rs1242761382 | 485 | N>T | No | gnomAD | |
| COSM4704683 | 486 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3513686 rs372242377 |
486 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs372242377 | 486 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1231178031 | 490 | Q>* | No |
TOPMed gnomAD |
|
|
RCV000596017 rs781085472 CA8388049 |
492 | F>missing | No |
ClinGen ClinVar dbSNP |
|
| rs1203067291 | 494 | H>Y | No |
TOPMed gnomAD |
|
| COSM6080169 | 495 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753280343 | 496 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs753280343 | 496 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs2072238137 | 497 | F>S | No | Ensembl | |
| rs538958497 | 498 | V>M | No | Ensembl | |
| rs1235454816 | 503 | E>* | No | TOPMed | |
| rs770798651 | 503 | E>D | No |
TOPMed gnomAD |
|
| rs192445309 | 504 | Y>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2142184730 | 505 | K>T | No | Ensembl | |
| rs1173798999 | 506 | K>* | No |
TOPMed gnomAD |
|
| rs746771415 | 507 | E>G | No | Ensembl | |
|
rs2072237590 COSM1179865 |
508 | G>D | prostate [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1402543930 | 508 | G>S | No |
TOPMed gnomAD |
|
| rs377600500 | 510 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs377600500 | 510 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1425743287 | 511 | W>* | No | gnomAD | |
| COSM6145648 | 511 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759294305 | 512 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs906879759 | 514 | I>V | No | Ensembl | |
| rs2072237208 | 516 | F>L | No | TOPMed | |
| rs2072237185 | 517 | G>E | No | Ensembl | |
| rs2072237154 | 518 | M>V | No | TOPMed | |
| rs763159867 | 521 | A>V | No |
ExAC gnomAD |
|
| rs1345983098 | 522 | A>G | No |
TOPMed gnomAD |
|
| rs769857967 | 522 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs748690722 | 524 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs748690722 | 524 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs2072237028 | 524 | I>V | No | Ensembl | |
| rs148038187 | 525 | E>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 525 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs148038187 | 525 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 525 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769185512 | 527 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs747362245 | 529 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs2142184636 | 529 | K>R | No | 1000Genomes | |
| rs777332782 | 530 | P>A | No |
ExAC gnomAD |
|
| TCGA novel | 530 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769279177 | 531 | L>V | No |
ExAC gnomAD |
|
| rs2142183600 | 532 | G>D | No | Ensembl | |
| rs747449994 | 533 | I>F | No |
ExAC gnomAD |
|
| rs775726283 | 534 | F>V | No | ExAC | |
| rs1046089949 | 536 | I>T | No | Ensembl | |
| rs1374469562 | 539 | E>G | No | gnomAD | |
| rs772433083 | 539 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs772433083 | 539 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs748923239 | 540 | E>K | No |
ExAC gnomAD |
|
| rs757657282 | 541 | C>F | No | gnomAD | |
| rs1445202853 | 541 | C>R | No | gnomAD | |
| rs757657282 | 541 | C>S | No | gnomAD | |
| COSM975215 | 542 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1218346694 COSM3513684 |
544 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs2072226812 | 544 | P>S | No | TOPMed | |
| rs991299916 | 545 | K>R | No |
TOPMed gnomAD |
|
| TCGA novel | 546 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs149040691 | 547 | T>K | No |
1000Genomes ESP ExAC gnomAD |
|
| TCGA novel | 548 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072226568 | 548 | D>N | No | TOPMed | |
| rs754915968 | 549 | T>N | No |
ExAC gnomAD |
|
| rs2072226434 | 550 | S>P | No | Ensembl | |
| TCGA novel | 551 | F>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766114551 | 551 | F>L | No |
ExAC gnomAD |
|
| rs758176237 | 553 | N>S | No |
ExAC gnomAD |
|
| rs750654484 | 555 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1481974689 | 555 | L>V | No |
TOPMed gnomAD |
|
|
COSM313040 rs762049118 |
556 | Y>* | lung [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs1380658768 RCV001171922 |
556 | Y>S | No |
ClinVar TOPMed dbSNP |
|
| rs1325602270 | 558 | Q>R | No | Ensembl | |
| rs761472735 | 560 | L>P | No | Ensembl | |
| rs546939198 | 560 | L>V | No |
1000Genomes TOPMed gnomAD |
|
| rs2072226003 | 561 | G>D | No | TOPMed | |
|
rs149876122 COSM110197 |
562 | K>N | skin [Cosmic] | No |
cosmic curated Ensembl |
| rs764209039 | 563 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs764209039 | 563 | S>F | No |
ExAC TOPMed gnomAD |
|
|
COSM3818783 rs201603486 |
564 | A>T | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
| rs772363050 | 564 | A>V | No |
ExAC gnomAD |
|
| rs111452886 | 565 | N>D | No |
TOPMed gnomAD |
|
| rs746219843 | 566 | F>C | No |
ExAC gnomAD |
|
| rs2072225712 | 567 | Q>* | No | Ensembl | |
| rs375076689 | 568 | K>R | No | Ensembl | |
| rs747734458 | 569 | P>A | No |
ExAC gnomAD |
|
| rs1385836788 | 570 | K>R | No |
TOPMed gnomAD |
|
| rs1385836788 | 570 | K>T | No |
TOPMed gnomAD |
|
| rs2072225536 | 571 | V>L | No | TOPMed | |
| rs780693693 | 572 | V>A | No |
ExAC gnomAD |
|
| rs754437166 | 573 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs550754244 | 576 | A>T | No |
ExAC TOPMed gnomAD |
|
|
COSM1380756 rs758179709 |
578 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1473802850 | 579 | H>R | No |
TOPMed gnomAD |
|
| rs1266587919 | 580 | F>L | No | gnomAD | |
| COSM4063805 | 580 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072225144 | 581 | S>C | No | gnomAD | |
| rs1007717319 | 581 | S>T | No | Ensembl | |
| rs201261875 | 582 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1338838301 | 583 | I>N | No | gnomAD | |
| rs140591842 | 585 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1363791536 | 586 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs754060887 | 587 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs764299420 | 589 | V>A | No |
ExAC gnomAD |
|
| rs764299420 | 589 | V>E | No |
ExAC gnomAD |
|
| rs2072224475 | 589 | V>M | No | gnomAD | |
| TCGA novel | 590 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1597402812 | 590 | D>Y | No | Ensembl | |
| rs1406583909 | 592 | N>S | No | gnomAD | |
| rs2072224171 | 596 | W>* | No | Ensembl | |
| rs1456340373 | 596 | W>* | No |
TOPMed gnomAD |
|
| rs753192400 | 596 | W>R | No |
ExAC gnomAD |
|
| TCGA novel | 597 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768054029 | 597 | L>P | No |
ExAC gnomAD |
|
| rs1597402797 | 598 | D>G | No | Ensembl | |
| COSM1479208 | 599 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2142183314 | 599 | K>N | No | Ensembl | |
| rs759886565 | 599 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs759886565 | 599 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs1477183477 | 600 | N>K | No | gnomAD | |
| rs201903868 | 601 | K>R | No | 1000Genomes | |
| rs1597402783 | 602 | D>A | No | Ensembl | |
| rs372583055 | 602 | D>N | No |
ESP TOPMed gnomAD |
|
| rs2072223824 | 603 | P>R | No | Ensembl | |
| rs1490962380 | 603 | P>S | No |
TOPMed gnomAD |
|
| rs934069206 | 605 | N>D | No |
TOPMed gnomAD |
|
| rs2072223631 | 606 | D>N | No |
TOPMed gnomAD |
|
| rs776229211 | 607 | T>A | No |
ExAC gnomAD |
|
| rs2072223537 | 608 | V>A | No | TOPMed | |
| rs746508341 | 609 | V>D | No |
ExAC TOPMed |
|
| rs768199968 | 609 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs768199968 | 609 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2072223418 | 610 | G>V | No | gnomAD | |
| rs1219378847 | 611 | L>P | No |
TOPMed gnomAD |
|
| rs772048786 | 612 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs745732819 | 614 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs745732819 | 614 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs1331836565 | 615 | S>P | No | gnomAD | |
| rs563022622 | 616 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs2072223063 | 617 | M>I | No | Ensembl | |
| rs757073823 | 617 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1420925722 | 617 | M>V | No |
TOPMed gnomAD |
|
| rs1194862920 | 618 | K>E | No | gnomAD | |
| COSM1479207 | 618 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754152665 | 619 | T>P | No |
ExAC gnomAD |
|
| rs754152665 | 619 | T>S | No |
ExAC gnomAD |
|
| rs1480568602 | 624 | F>S | No | gnomAD | |
| rs756204294 | 625 | S>A | No |
ExAC gnomAD |
|
| rs541931160 | 625 | S>C | No |
1000Genomes TOPMed gnomAD |
|
| rs541931160 | 625 | S>F | No |
1000Genomes TOPMed gnomAD |
|
| rs541931160 | 625 | S>Y | No |
1000Genomes TOPMed gnomAD |
|
| rs574510840 | 626 | T>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs574510840 | 626 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs752816791 | 626 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs183183083 | 627 | Y>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs752012893 | 627 | Y>C | No |
ExAC gnomAD |
|
| rs752012893 | 627 | Y>F | No |
ExAC gnomAD |
|
| rs2072222512 | 628 | A>G | No | TOPMed | |
| rs763275859 | 628 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2072222432 | 629 | S>G | No | TOPMed | |
| rs2072222408 | 630 | A>P | No |
TOPMed gnomAD |
|
| rs148707844 | 633 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs758822435 | 633 | D>G | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 633 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758822435 | 633 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs765459916 | 635 | S>N | No |
ExAC gnomAD |
|
|
COSM2924775 rs529314902 |
636 | A>T | pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs774487685 | 639 | G>R | No |
ExAC gnomAD |
|
| rs557143201 | 639 | G>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1597402655 | 640 | A>P | No | Ensembl | |
| rs2072220924 | 640 | A>V | No | TOPMed | |
| TCGA novel | 641 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1185937455 | 641 | K>T | No | gnomAD | |
| rs2072220843 | 642 | K>T | No | TOPMed | |
| rs749169527 | 643 | K>N | No |
ExAC gnomAD |
|
| rs1236963519 | 644 | G>S | No | gnomAD | |
| rs1185439043 | 645 | S>T | No |
TOPMed gnomAD |
|
| rs535957016 | 646 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA |
| rs773075395 | 648 | Q>E | No |
ExAC gnomAD |
|
| TCGA novel | 649 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781240971 | 650 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs781240971 | 650 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs780607733 | 652 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1225738007 | 653 | L>F | No | TOPMed | |
| rs1225738007 | 653 | L>I | No | TOPMed | |
| COSM3513680 | 656 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072200668 | 662 | M>T | No | Ensembl | |
| rs1749995981 | 663 | T>A | No | Ensembl | |
| rs139649943 | 663 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs139649943 | 663 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs754146272 | 665 | L>V | No |
ExAC gnomAD |
|
| rs1036689482 | 666 | R>K | No |
TOPMed gnomAD |
|
| rs778132480 | 666 | R>S | No |
ExAC gnomAD |
|
| rs1567686350 | 668 | T>R | No | Ensembl | |
| rs2072200353 | 669 | H>Y | No |
TOPMed gnomAD |
|
| rs1424405891 | 670 | P>S | No |
TOPMed gnomAD |
|
| rs1424405891 | 670 | P>T | No |
TOPMed gnomAD |
|
| COSM3513679 | 671 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1169981995 COSM559504 COSM6080170 |
671 | H>Y | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs12936716 | 672 | F>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM1128833 | 673 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs142181278 COSM975213 |
673 | V>I | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs762523289 | 674 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs530070079 | 675 | C>Y | No |
1000Genomes ExAC gnomAD |
|
| rs776298958 | 676 | I>N | No |
ExAC gnomAD |
|
| rs761737413 | 676 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1256890042 | 677 | I>L | No | gnomAD | |
| TCGA novel | 677 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768386064 | 679 | N>S | No |
1000Genomes ExAC gnomAD |
|
| COSM3513678 | 680 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775653821 | 683 | T>A | No |
ExAC TOPMed gnomAD |
|
|
rs772185130 COSM313039 |
683 | T>I | lung [Cosmic] | No |
cosmic curated ExAC |
| COSM3889215 | 684 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1749852 | 685 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139004551 | 685 | G>W | No |
ESP ExAC TOPMed gnomAD |
|
| COSM283035 | 686 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1303702809 | 688 | E>Q | No | gnomAD | |
| rs1465743262 | 689 | H>N | No | gnomAD | |
| rs760438917 | 690 | E>K | No |
ExAC gnomAD |
|
| rs2072198357 | 691 | L>P | No | gnomAD | |
|
rs775743614 COSM975211 |
692 | V>A | endometrium [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1381616912 | 692 | V>L | No |
TOPMed gnomAD |
|
| rs1425096175 | 694 | H>Y | No | gnomAD | |
| rs1256256315 | 695 | Q>* | No | gnomAD | |
| rs1180699176 | 695 | Q>H | No | gnomAD | |
| rs2072198073 | 696 | L>M | No | TOPMed | |
| rs369392212 | 696 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072198013 | 697 | R>K | No | Ensembl | |
| rs145148429 | 697 | R>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1264819393 | 699 | N>D | No | gnomAD | |
| rs957536307 | 700 | G>C | No |
TOPMed gnomAD |
|
| rs774308064 | 701 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1158371179 | 703 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1286014108 | 704 | G>A | No | gnomAD | |
| rs1286014108 | 704 | G>V | No | gnomAD | |
| rs770992448 | 705 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs773825015 | 706 | R>C | No |
ExAC TOPMed gnomAD |
|
| COSM417208 | 708 | C>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1300256966 | 708 | C>Y | No | gnomAD | |
|
rs1060499731 RCV000596376 CA16609482 |
710 | K>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
COSM107553 rs141489827 |
711 | G>E | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs373333893 | 712 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| COSM975210 | 713 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 714 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs371932735 | 714 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 715 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072197342 | 715 | R>S | No | TOPMed | |
|
RCV000239061 RCV000897380 rs115483891 CA8387852 |
716 | I>N | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs745471357 | 716 | I>V | No |
ExAC gnomAD |
|
| rs1425377153 | 718 | Y>C | No | gnomAD | |
| rs201064047 | 718 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs201064047 | 718 | Y>N | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 719 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072197160 | 719 | G>V | No | TOPMed | |
| rs1193930864 | 721 | F>Y | No | gnomAD | |
| rs764003729 | 722 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs755968058 | 723 | Q>K | No |
ExAC gnomAD |
|
| rs1159067453 | 725 | Y>* | No |
TOPMed gnomAD |
|
|
rs565515290 RCV000893860 |
725 | Y>* | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1008433257 | 725 | Y>H | No | TOPMed | |
| rs2072193729 | 726 | K>E | No | Ensembl | |
| COSM6145649 | 726 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1359689882 | 726 | K>R | No |
TOPMed gnomAD |
|
| rs1317539434 | 727 | V>D | No |
TOPMed gnomAD |
|
| rs756057621 | 727 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs756057621 | 727 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2142180261 | 728 | L>S | No | Ensembl | |
| rs752582352 | 729 | N>I | No |
ExAC gnomAD |
|
| rs2072193484 | 730 | A>G | No | Ensembl | |
| rs2072193506 | 730 | A>T | No | TOPMed | |
| rs767246642 | 731 | S>R | No |
ExAC gnomAD |
|
| rs2072193367 | 734 | P>Q | No | TOPMed | |
| rs1438643927 | 735 | E>G | No | gnomAD | |
| rs2072193302 | 736 | G>A | No |
TOPMed gnomAD |
|
| rs754772545 | 736 | G>R | No |
ExAC gnomAD |
|
| rs2072193252 | 737 | Q>R | No | Ensembl | |
| rs1435208666 | 738 | F>L | No | TOPMed | |
| rs766612483 | 738 | F>L | No |
ExAC gnomAD |
|
| rs766612483 | 738 | F>V | No |
ExAC gnomAD |
|
| rs996857100 | 739 | I>T | No | TOPMed | |
| rs763011772 | 741 | S>N | No |
ExAC gnomAD |
|
| COSM6145650 | 741 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM6145651 | 742 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072193064 | 744 | A>P | No | Ensembl | |
|
RCV001765467 rs2072193064 |
744 | A>T | No |
ClinVar Ensembl dbSNP |
|
|
CA398118864 RCV000522484 rs1555556803 |
745 | S>F | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs2072192966 | 746 | E>K | No | Ensembl | |
| rs1300603955 | 747 | K>N | No |
TOPMed gnomAD |
|
| rs1378585240 | 748 | L>F | No | gnomAD | |
| TCGA novel | 748 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765353703 | 750 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs1331689301 | 750 | A>T | No | gnomAD | |
| rs765353703 | 750 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2142180192 | 752 | I>T | No | Ensembl | |
| rs762437428 | 752 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1043670011 | 753 | D>A | No |
TOPMed gnomAD |
|
| rs1043670011 | 753 | D>G | No |
TOPMed gnomAD |
|
| rs1043670011 | 753 | D>V | No |
TOPMed gnomAD |
|
| rs1353450116 | 754 | I>V | No |
TOPMed gnomAD |
|
| rs560062462 | 756 | H>D | No | 1000Genomes | |
| COSM1302398 | 756 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1271227321 | 757 | T>I | No | gnomAD | |
| rs1416116213 | 758 | Q>E | No | gnomAD | |
| rs945295096 | 758 | Q>P | No | Ensembl | |
| rs945295096 | 758 | Q>R | No | Ensembl | |
| rs773940118 | 759 | Y>H | No |
ExAC gnomAD |
|
| TCGA novel | 764 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2142180154 | 765 | K>* | No | Ensembl | |
| rs1310664814 | 768 | F>I | No |
TOPMed gnomAD |
|
| rs1310664814 | 768 | F>L | No |
TOPMed gnomAD |
|
| rs775934311 | 768 | F>L | No |
ExAC gnomAD |
|
| TCGA novel | 769 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3969797 | 770 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs944808986 | 771 | G>R | No | TOPMed | |
| rs772275170 | 772 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs762623757 | 772 | L>P | No |
ExAC gnomAD |
|
| rs772819714 | 773 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1400643420 | 773 | L>V | No | gnomAD | |
| COSM4141971 | 774 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1473154986 | 777 | E>* | No | gnomAD | |
| COSM3988755 | 777 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1473154986 | 777 | E>K | No | gnomAD | |
| rs769244945 | 778 | E>G | No |
ExAC TOPMed gnomAD |
|
| COSM4593875 | 778 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1186075035 | 779 | M>I | No | gnomAD | |
| rs1597401380 | 779 | M>T | No | Ensembl | |
| rs1489183481 | 781 | D>G | No | gnomAD | |
| rs1459149569 | 781 | D>H | No |
TOPMed gnomAD |
|
|
rs781058661 COSM223851 |
782 | E>K | skin [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1291904253 | 784 | L>F | No | gnomAD | |
| rs1445231364 | 784 | L>S | No | gnomAD | |
| TCGA novel | 785 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072190329 | 788 | I>L | No | TOPMed | |
| rs1228434133 | 788 | I>T | No | gnomAD | |
| rs2072190329 | 788 | I>V | No | TOPMed | |
| rs1355031105 | 789 | T>R | No | gnomAD | |
| COSM1380754 | 790 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072190174 | 791 | T>R | No | Ensembl | |
| rs768644838 | 791 | T>S | No |
ExAC gnomAD |
|
| rs746866721 | 792 | Q>P | No |
ExAC gnomAD |
|
| rs746866721 | 792 | Q>R | No |
ExAC gnomAD |
|
| rs779923682 | 793 | A>D | No |
ExAC gnomAD |
|
| TCGA novel | 794 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072190009 | 795 | C>R | No | Ensembl | |
| rs1214736532 | 796 | R>S | No | gnomAD | |
| rs758230635 | 797 | G>A | No |
ExAC gnomAD |
|
| rs758230635 | 797 | G>E | No |
ExAC gnomAD |
|
| rs142030000 | 798 | F>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1368868765 | 799 | L>Q | No |
TOPMed gnomAD |
|
| rs1162716520 | 800 | M>I | No |
TOPMed gnomAD |
|
| rs1472316809 | 800 | M>V | No |
TOPMed gnomAD |
|
| rs1326077263 | 801 | R>T | No | gnomAD | |
| rs2072189554 | 802 | V>A | No | TOPMed | |
| rs757475660 | 802 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs753954757 | 803 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs753954757 | 803 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs866672047 | 805 | Q>* | No | Ensembl | |
| TCGA novel | 806 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM705173 rs962009902 |
807 | M>I | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated Ensembl NCI-TCGA Cosmic |
| rs2072189373 | 808 | L>S | No | TOPMed | |
| rs2072189373 | 808 | L>W | No | TOPMed | |
| rs1015870097 | 809 | Q>E | No |
TOPMed gnomAD |
|
|
rs1478940960 COSM3513675 |
810 | R>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| COSM3513676 | 810 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072189293 | 811 | R>T | No | Ensembl | |
| TCGA novel | 812 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772047264 | 812 | E>D | No |
ExAC TOPMed |
|
| rs1231873171 | 812 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 813 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745650365 | 814 | L>F | No |
ExAC gnomAD |
|
| rs757564301 | 815 | F>C | No |
ExAC TOPMed gnomAD |
|
| rs778780905 | 815 | F>L | No |
ExAC gnomAD |
|
| rs2072173290 | 816 | C>R | No | gnomAD | |
| TCGA novel | 818 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs143710889 | 819 | Y>H | No |
1000Genomes ExAC gnomAD |
|
| rs2072173174 | 820 | N>D | No | TOPMed | |
| TCGA novel | 820 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1457798399 | 821 | V>F | No | gnomAD | |
| rs1457798399 | 821 | V>I | No | gnomAD | |
| rs1257110531 | 822 | R>C | No |
TOPMed gnomAD |
|
|
RCV001354687 rs776002223 |
822 | R>H | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| COSM6145652 | 822 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs373659738 | 823 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4141970 rs1004863060 |
823 | A>T | ovary [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs755403538 | 824 | F>L | No |
ExAC TOPMed |
|
| rs1419994195 | 825 | M>I | No | TOPMed | |
| rs751927704 | 825 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs770622195 | 826 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs867870408 | 827 | V>G | No |
TOPMed gnomAD |
|
| rs761551580 | 827 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs761551580 | 827 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs369293931 | 829 | H>Y | No |
ESP TOPMed gnomAD |
|
| rs1440295473 | 830 | W>C | No | gnomAD | |
| rs763703050 | 830 | W>G | No |
ExAC gnomAD |
|
| rs2072172312 | 831 | P>L | No | Ensembl | |
| rs746586658 | 832 | W>* | No | Ensembl | |
| rs2142178427 | 832 | W>C | No | Ensembl | |
|
COSM1709827 COSM1380753 rs866239257 |
833 | M>I | large_intestine skin [Cosmic] | No |
cosmic curated Ensembl |
| rs2072172156 | 834 | K>I | No | Ensembl | |
| rs2072172118 | 838 | K>R | No | gnomAD | |
| rs1361071176 | 839 | I>T | No |
TOPMed gnomAD |
|
| TCGA novel | 840 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1176552857 | 841 | P>A | No | gnomAD | |
| rs995598504 | 843 | L>F | No |
TOPMed gnomAD |
|
| COSM975208 | 843 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775630833 | 844 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs1439837914 | 845 | S>G | No | gnomAD | |
| rs772137170 | 848 | T>A | No |
ExAC gnomAD |
|
| COSM705175 | 849 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774307650 | 849 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs770808549 | 851 | E>D | No |
ExAC TOPMed gnomAD |
|
| COSM1479206 | 851 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV000733690 rs1319449107 COSM705176 |
853 | A>T | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ClinVar NCI-TCGA dbSNP gnomAD |
| rs777836357 | 854 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs777836357 | 854 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1567685307 | 855 | M>K | No | Ensembl | |
| rs756275515 | 855 | M>L | No |
ExAC gnomAD |
|
| COSM3513672 | 857 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781182860 | 858 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs1233031989 | 858 | E>A | No |
TOPMed gnomAD |
|
| rs781182860 | 858 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs543847124 | 860 | Q>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs377068313 | 861 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072170675 | 861 | K>N | No |
TOPMed gnomAD |
|
| rs2072170641 | 862 | T>I | No | Ensembl | |
| rs758662426 | 863 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 864 | D>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs943155213 | 865 | E>D | No | TOPMed | |
| rs1011679307 | 867 | A>G | No |
TOPMed gnomAD |
|
| rs774805115 | 867 | A>T | No |
ExAC TOPMed gnomAD |
|
|
rs138755767 COSM975207 |
873 | R>Q | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs763645108 | 873 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs376614320 | 875 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072169983 | 877 | E>G | No |
TOPMed gnomAD |
|
| COSM705177 | 877 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1219195879 | 878 | E>D | No | gnomAD | |
| rs759575082 | 878 | E>G | No |
ExAC gnomAD |
|
| COSM3513670 | 878 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1355321608 | 879 | K>E | No |
TOPMed gnomAD |
|
|
COSM51304 rs774397725 COSM1380752 |
880 | M>I | large_intestine breast [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs770808142 | 881 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1357082304 | 882 | T>S | No |
TOPMed gnomAD |
|
| rs1296205510 | 882 | T>S | No | TOPMed | |
| rs2072169476 | 883 | L>I | No |
TOPMed gnomAD |
|
| TCGA novel | 885 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1423076367 | 886 | E>D | No | gnomAD | |
|
rs1294198951 COSM3513669 |
886 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs189744808 | 888 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1384210757 | 891 | Q>K | No | gnomAD | |
| rs2072169123 | 893 | Q>P | No | gnomAD | |
| rs35952774 | 894 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199556347 | 896 | S>C | No | Ensembl | |
|
COSM3513667 rs1256915000 |
897 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs201276413 | 898 | A>T | No |
TOPMed gnomAD |
|
| COSM3513666 | 899 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1443638268 | 899 | D>Y | No | Ensembl | |
| TCGA novel | 902 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs62058112 | 904 | A>G | No | Ensembl | |
| rs776805911 | 904 | A>T | No |
ExAC gnomAD |
|
| rs768776876 | 905 | E>D | No |
ExAC gnomAD |
|
| rs1435459884 | 907 | R>G | No | gnomAD | |
| rs2072145443 | 907 | R>M | No | TOPMed | |
| rs568395488 | 909 | E>K | No | 1000Genomes | |
|
rs1254125426 COSM3957976 |
910 | Q>* | lung [Cosmic] | No |
cosmic curated gnomAD |
| rs1193146422 | 910 | Q>P | No | gnomAD | |
| TCGA novel | 911 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780682650 | 914 | N>T | No |
ExAC TOPMed gnomAD |
|
|
RCV000595771 rs1555556193 CA16609481 |
916 | I>missing | No |
ClinGen ClinVar dbSNP |
|
| rs772036029 | 918 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs746012879 | 920 | A>D | No |
ExAC gnomAD |
|
| rs1448637732 | 920 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 920 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746012879 | 920 | A>V | No |
ExAC gnomAD |
|
| rs1235076936 | 921 | K>R | No |
TOPMed gnomAD |
|
| TCGA novel | 922 | I>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_030207 rs4372733 |
924 | E>G | No |
UniProt Ensembl dbSNP |
|
| rs757532571 | 925 | V>M | No |
ExAC gnomAD |
|
| rs1363574693 | 926 | T>A | No | gnomAD | |
| rs571369477 | 926 | T>N | No |
ExAC TOPMed gnomAD |
|
|
rs781149673 RCV000996489 |
927 | E>Q | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs2072144772 COSM357253 |
930 | E>Q | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs1393822149 | 931 | E>D | No | gnomAD | |
|
COSM3513665 rs1166542049 |
932 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| COSM4063803 | 933 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs147618682 | 934 | E>D | No |
ESP TOPMed gnomAD |
|
| COSM4837152 | 935 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1320731738 | 935 | I>V | No | TOPMed | |
| rs757824042 | 936 | N>S | No |
ExAC TOPMed |
|
| rs149946448 | 937 | A>G | No |
ESP TOPMed gnomAD |
|
| rs2072144384 | 937 | A>S | No | TOPMed | |
| rs149946448 | 937 | A>V | No |
ESP TOPMed gnomAD |
|
| rs764753631 | 938 | E>* | No |
ExAC gnomAD |
|
| rs1268984917 | 941 | A>V | No | Ensembl | |
| rs776918994 | 942 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs761474736 | 942 | K>R | No |
ExAC gnomAD |
|
| rs764443529 | 943 | K>M | No |
ExAC gnomAD |
|
| COSM1380751 | 943 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764443529 | 943 | K>T | No |
ExAC gnomAD |
|
| rs1788738157 | 944 | R>I | No | Ensembl | |
| rs761089254 | 946 | L>P | No |
ExAC gnomAD |
|
| rs775842493 | 948 | D>N | No |
ExAC gnomAD |
|
| rs772677984 | 948 | D>V | No |
ExAC gnomAD |
|
| rs551907964 | 950 | C>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2142176046 | 951 | S>* | No | Ensembl | |
| COSM4892757 | 951 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM975205 | 954 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1597399789 | 954 | K>R | No | Ensembl | |
| COSM3513664 | 956 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs771221561 | 957 | I>T | No |
ExAC gnomAD |
|
| rs1232009976 | 957 | I>V | No |
TOPMed gnomAD |
|
| rs749525882 | 958 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs2072143681 | 959 | D>E | No | gnomAD | |
| rs1333674216 | 959 | D>Y | No | gnomAD | |
| rs2072143639 | 960 | L>F | No | TOPMed | |
| COSM2924758 | 960 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1248202086 | 961 | E>Q | No | TOPMed | |
| rs754806719 | 962 | L>M | No |
ExAC gnomAD |
|
| TCGA novel | 963 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1417069852 | 963 | T>I | No | gnomAD | |
| rs2072143524 | 965 | A>P | No | Ensembl | |
| rs779823632 | 966 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs758389483 | 967 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs758389483 | 967 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1433432122 | 969 | K>Q | No | gnomAD | |
| rs1140933 | 970 | E>* | No |
ExAC gnomAD |
|
|
rs1140933 COSM3513663 |
970 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1140933 | 970 | E>Q | No |
ExAC gnomAD |
|
| rs756832002 | 971 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1469662832 | 972 | H>Y | No | gnomAD | |
| rs753480257 | 973 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs753480257 | 973 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs753480257 | 973 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs151035575 | 974 | T>A | No | ESP | |
|
COSM472268 rs763789155 |
974 | T>M | kidney [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| COSM435985 | 974 | T>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3513662 | 978 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1567684297 | 978 | V>A | No | Ensembl | |
| rs1164569278 | 978 | V>M | No |
TOPMed gnomAD |
|
| rs1213718917 | 980 | N>T | No |
TOPMed gnomAD |
|
| rs2072141242 | 981 | L>H | No | Ensembl | |
| TCGA novel | 983 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1406305642 | 986 | A>P | No |
TOPMed gnomAD |
|
| rs1406305642 | 986 | A>T | No |
TOPMed gnomAD |
|
| rs752949209 | 989 | D>E | No |
ExAC gnomAD |
|
| rs1271068676 | 991 | T>I | No | gnomAD | |
| rs767929562 | 992 | I>T | No |
ExAC gnomAD |
|
| rs2072140985 | 992 | I>V | No | Ensembl | |
| COSM3513661 | 993 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759884889 | 993 | A>V | No |
ExAC gnomAD |
|
| rs992750152 | 994 | K>N | No | Ensembl | |
|
TCGA novel rs75477725 |
996 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA 1000Genomes ExAC TOPMed gnomAD |
| rs2072140712 | 996 | S>T | No | Ensembl | |
| rs1394082972 | 997 | K>E | No | gnomAD | |
| rs1034267182 | 997 | K>R | No | Ensembl | |
|
rs763093580 COSM559507 |
998 | E>Q | lung [Cosmic] | No |
cosmic curated ExAC |
| rs1411515344 | 999 | K>R | No |
TOPMed gnomAD |
|
| rs1411515344 | 999 | K>T | No |
TOPMed gnomAD |
|
|
rs147185960 RCV000931294 |
1000 | K>E | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs775077561 | 1000 | K>N | No |
ExAC TOPMed gnomAD |
|
|
rs143531631 RCV000931293 |
1000 | K>T | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1237160219 | 1001 | A>G | No |
TOPMed gnomAD |
|
| rs139605305 | 1002 | L>F | No |
ESP TOPMed gnomAD |
|
| rs2072140121 | 1004 | E>D | No | Ensembl | |
| rs2072140140 | 1004 | E>K | No | TOPMed | |
| rs2072140140 | 1004 | E>Q | No | TOPMed | |
| rs2072140087 | 1005 | T>A | No | Ensembl | |
| TCGA novel | 1006 | H>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA207012 rs750008371 RCV000193490 |
1007 | Q>missing | No |
ClinGen ClinVar dbSNP |
|
| rs1193775043 | 1007 | Q>E | No | gnomAD | |
| TCGA novel | 1007 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745704153 | 1008 | Q>* | No |
ExAC gnomAD |
|
| rs1307657300 | 1008 | Q>R | No | gnomAD | |
| rs778795494 | 1009 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs770889098 | 1010 | L>R | No |
ExAC gnomAD |
|
| TCGA novel | 1011 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1306236434 | 1012 | D>E | No | gnomAD | |
| rs368176331 | 1015 | A>T | No |
ESP ExAC |
|
| COSM4063802 | 1016 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1346159144 | 1016 | E>K | No |
TOPMed gnomAD |
|
| COSM6145653 | 1017 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs146341182 | 1017 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1205708349 | 1018 | D>G | No |
TOPMed gnomAD |
|
| rs755657441 | 1018 | D>N | No |
ExAC gnomAD |
|
| COSM3402579 | 1019 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072139477 | 1019 | K>N | No | TOPMed | |
| rs1567684226 | 1020 | V>I | No | TOPMed | |
| rs1444669759 | 1021 | N>H | No |
TOPMed gnomAD |
|
| rs752390040 | 1022 | I>T | No |
ExAC gnomAD |
|
| rs755347864 | 1026 | A>G | No |
ExAC gnomAD |
|
| rs755347864 | 1026 | A>V | No |
ExAC gnomAD |
|
| rs563278731 | 1029 | K>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs374267154 | 1030 | L>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072139162 | 1030 | L>V | No | gnomAD | |
| rs1453653455 | 1031 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1032 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072139032 | 1033 | Q>* | No | TOPMed | |
| rs200190381 | 1034 | V>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs750470825 | 1035 | D>N | No |
ExAC TOPMed gnomAD |
|
|
COSM1493789 rs2072138923 |
1036 | D>G | kidney [Cosmic] | No |
cosmic curated Ensembl |
| rs2142175499 | 1037 | L>V | No | 1000Genomes | |
| rs2072137729 | 1042 | E>G | No | Ensembl | |
| rs540108157 | 1042 | E>Q | No |
ExAC gnomAD |
|
| COSM3513659 | 1044 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs762795207 | 1045 | K>R | No |
ExAC gnomAD |
|
| rs941141027 | 1046 | K>N | No | TOPMed | |
| rs201501689 | 1046 | K>Q | No | Ensembl | |
| rs1036720263 | 1046 | K>T | No |
TOPMed gnomAD |
|
| rs2072137443 | 1047 | L>H | No | Ensembl | |
| rs2072137471 | 1047 | L>V | No | TOPMed | |
|
rs977910326 COSM975203 |
1048 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs977910326 | 1048 | R>G | No |
TOPMed gnomAD |
|
|
COSM180552 rs773112769 |
1048 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs201992379 | 1049 | M>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1290543576 | 1050 | D>A | No |
TOPMed gnomAD |
|
| rs1290543576 | 1050 | D>V | No |
TOPMed gnomAD |
|
| rs1156239645 | 1052 | E>G | No | gnomAD | |
| rs1462722902 | 1053 | R>G | No | gnomAD | |
|
rs2072137082 COSM3513658 |
1053 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| rs140112749 | 1056 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM135740 rs140112749 |
1056 | R>Q | ovary Variant assessed as Somatic; MODERATE impact. skin [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1189159212 | 1056 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2072136859 | 1057 | K>R | No | TOPMed | |
| rs1188806237 | 1058 | L>Q | No | gnomAD | |
| rs2072136837 | 1058 | L>V | No | TOPMed | |
| TCGA novel | 1059 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1034707927 | 1060 | G>D | No | gnomAD | |
| rs2072136783 | 1060 | G>R | No | TOPMed | |
| rs1034707927 | 1060 | G>V | No | gnomAD | |
| rs746661016 | 1061 | D>Y | No |
ExAC gnomAD |
|
| rs2142175383 | 1062 | L>R | No | Ensembl | |
| rs758818675 | 1064 | L>S | No |
ExAC gnomAD |
|
| rs779438748 | 1065 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1338477841 | 1065 | A>V | No | gnomAD | |
| rs2072136397 | 1066 | Q>* | No | gnomAD | |
| TCGA novel | 1067 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1297364390 | 1068 | S>C | No | gnomAD | |
|
rs1297364390 COSM3513657 |
1068 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs757730518 | 1069 | T>A | No |
ExAC TOPMed gnomAD |
|
| COSM4837307 | 1069 | T>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072136277 | 1070 | M>T | No | gnomAD | |
| rs753899429 | 1070 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1201459194 | 1071 | D>G | No |
TOPMed gnomAD |
|
| COSM3513656 | 1071 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764088983 | 1071 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs2072136171 | 1072 | M>T | No | TOPMed | |
| rs952671809 | 1072 | M>V | No | Ensembl | |
| TCGA novel | 1073 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs867392074 | 1073 | E>K | No | Ensembl | |
| rs2072136080 | 1075 | D>Y | No | TOPMed | |
| rs2072136045 | 1077 | Q>E | No | gnomAD | |
| rs149628973 | 1079 | L>F | No |
ESP TOPMed gnomAD |
|
| rs752905198 | 1080 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs756254146 | 1080 | D>N | No |
ExAC gnomAD |
|
| rs2072135852 | 1082 | K>Q | No | Ensembl | |
| rs2072135825 | 1083 | L>I | No | Ensembl | |
| TCGA novel | 1084 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3513655 | 1085 | K>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764945421 | 1085 | K>N | No |
ExAC gnomAD |
|
| COSM975202 | 1087 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs761731391 | 1087 | E>K | No |
ExAC gnomAD |
|
| rs761731391 | 1087 | E>Q | No |
ExAC gnomAD |
|
| COSM3513654 | 1089 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764172364 | 1091 | S>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1091 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs917379039 | 1092 | N>S | No |
TOPMed gnomAD |
|
| rs1270201653 | 1093 | L>V | No |
TOPMed gnomAD |
|
| rs1252907424 | 1095 | S>C | No |
TOPMed gnomAD |
|
|
COSM240793 rs760227712 |
1097 | I>T | prostate [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs775166125 | 1099 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1197400289 | 1100 | E>G | No |
TOPMed gnomAD |
|
| rs771797841 | 1100 | E>K | No |
ExAC gnomAD |
|
| rs1197400289 | 1100 | E>V | No |
TOPMed gnomAD |
|
| COSM435984 | 1101 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759117723 | 1103 | V>A | No |
ExAC gnomAD |
|
|
rs150717297 COSM975201 |
1104 | E>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs749713546 | 1106 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs749713546 | 1106 | Q>E | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1106 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2142175122 | 1107 | L>I | No | Ensembl | |
| rs1307243207 | 1108 | Q>* | No | gnomAD | |
| TCGA novel | 1109 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1392400393 | 1109 | K>N | No |
TOPMed gnomAD |
|
| rs1370339539 | 1110 | K>T | No | gnomAD | |
| rs778111589 | 1111 | I>F | No |
ExAC gnomAD |
|
| rs1555555996 | 1111 | I>T | No | Ensembl | |
| rs778111589 | 1111 | I>V | No |
ExAC gnomAD |
|
| TCGA novel | 1113 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770318554 | 1113 | E>G | No |
ExAC gnomAD |
|
|
TCGA novel rs2072133821 |
1113 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs755125452 | 1115 | Q>* | No |
ExAC gnomAD |
|
| rs755125452 | 1115 | Q>E | No |
ExAC gnomAD |
|
| rs972563832 | 1116 | A>D | No | gnomAD | |
|
COSM559508 rs2072132763 COSM6080173 |
1117 | R>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic Ensembl |
| rs1387251939 | 1117 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1387251939 | 1117 | R>L | No | gnomAD | |
| rs2142174971 | 1118 | I>F | No | Ensembl | |
| rs373569106 | 1118 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| COSM975200 | 1119 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768705671 | 1121 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs2072132615 | 1121 | L>P | No | Ensembl | |
| rs747160540 | 1122 | G>R | No |
ExAC gnomAD |
|
| TCGA novel | 1123 | E>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs780104656 COSM3513652 |
1123 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1256801519 | 1125 | I>L | No | gnomAD | |
|
rs748982401 COSM975199 |
1126 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2072132335 | 1126 | E>V | No | Ensembl | |
| rs2072132309 | 1127 | A>V | No | TOPMed | |
| rs2072132280 | 1128 | E>* | No | gnomAD | |
| rs777719193 | 1128 | E>A | No |
ExAC gnomAD |
|
| TCGA novel | 1128 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1128 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1203991660 | 1129 | R>G | No | gnomAD | |
| rs1164016558 | 1130 | A>E | No |
TOPMed gnomAD |
|
| rs373278832 | 1131 | S>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs1335919613 | 1131 | S>P | No | gnomAD | |
| rs754417813 | 1132 | R>* | No |
ExAC gnomAD |
|
|
COSM3387699 rs1459405999 |
1132 | R>Q | pancreas [Cosmic] | No |
cosmic curated TOPMed gnomAD |
|
COSM3937236 rs936506457 |
1133 | A>T | oesophagus [Cosmic] | No |
cosmic curated TOPMed |
| rs1440801997 | 1133 | A>V | No |
TOPMed gnomAD |
|
| COSM3513651 | 1136 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs529877880 | 1139 | R>G | No |
1000Genomes ExAC gnomAD |
|
|
COSM3969796 rs2072131649 |
1139 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs773499534 | 1141 | D>N | No |
ExAC gnomAD |
|
| rs773499534 | 1141 | D>Y | No |
ExAC gnomAD |
|
| COSM975198 | 1142 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765579840 | 1143 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs1161189464 | 1144 | R>G | No |
TOPMed gnomAD |
|
|
COSM2924742 rs1382406133 |
1144 | R>Q | pancreas [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1161189464 | 1144 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1476366790 | 1145 | E>K | No |
TOPMed gnomAD |
|
| rs762222027 | 1146 | L>R | No |
ExAC gnomAD |
|
| rs2072131155 | 1147 | E>Q | No | TOPMed | |
| rs1191926786 | 1148 | E>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3513649 | 1150 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs769243679 | 1150 | S>R | No | ExAC | |
| rs777096656 | 1150 | S>R | No |
ExAC gnomAD |
|
| rs2072130965 | 1151 | E>D | No | TOPMed | |
| rs1254267380 | 1152 | R>K | No |
TOPMed gnomAD |
|
| rs1254267380 | 1152 | R>T | No |
TOPMed gnomAD |
|
| TCGA novel | 1153 | L>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747018022 | 1157 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1567683756 | 1158 | G>A | No | Ensembl | |
| rs1773358432 | 1158 | G>R | No |
TOPMed gnomAD |
|
| rs1567683756 | 1158 | G>V | No | Ensembl | |
| rs1597399223 | 1159 | A>G | No | Ensembl | |
| TCGA novel | 1159 | A>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4063801 | 1160 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2142174812 | 1161 | S>A | No | Ensembl | |
| COSM435983 | 1161 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1226935824 | 1162 | A>D | No |
TOPMed gnomAD |
|
| rs1289655596 | 1162 | A>P | No | gnomAD | |
| rs1242309135 | 1163 | Q>* | No |
TOPMed gnomAD |
|
| rs1271271357 | 1164 | V>G | No | gnomAD | |
| rs180922386 | 1164 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1188612954 | 1167 | N>K | No |
TOPMed gnomAD |
|
| rs2072130201 | 1168 | K>R | No | TOPMed | |
| rs746087794 | 1170 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM435982 rs2072130162 |
1170 | R>W | breast [Cosmic] | No |
cosmic curated gnomAD |
| TCGA novel | 1171 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1420099206 | 1171 | E>Q | No | Ensembl | |
| rs1265311251 | 1172 | A>V | No | gnomAD | |
| TCGA novel | 1173 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1175 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1441942691 | 1175 | Q>R | No |
TOPMed gnomAD |
|
| rs111350680 | 1177 | L>Q | No | Ensembl | |
| rs1321385050 | 1178 | R>H | No |
TOPMed gnomAD |
|
| rs1321385050 | 1178 | R>L | No |
TOPMed gnomAD |
|
| rs142073810 | 1178 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1411423641 | 1180 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1411423641 | 1180 | D>N | No |
TOPMed gnomAD |
|
| rs1450883772 | 1181 | L>P | No | TOPMed | |
| COSM6080174 | 1184 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372242216 | 1184 | A>S | No |
ESP TOPMed |
|
| rs2072129389 | 1184 | A>V | No | TOPMed | |
| TCGA novel | 1185 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs138064925 | 1185 | T>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs138064925 RCV000912880 |
1185 | T>S | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs754967636 | 1187 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs754967636 | 1187 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1006354139 | 1188 | H>R | No |
TOPMed gnomAD |
|
| rs2072128948 | 1189 | E>V | No |
TOPMed gnomAD |
|
| rs1219898616 | 1191 | M>I | No | gnomAD | |
| rs889294666 | 1192 | V>A | No | TOPMed | |
| COSM6080175 | 1193 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1274073145 | 1194 | A>P | No | gnomAD | |
|
rs1340594495 COSM4704678 |
1196 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM3889212 rs765916450 |
1196 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs757912324 | 1197 | K>N | No |
ExAC gnomAD |
|
| rs2072128582 | 1198 | K>T | No | Ensembl | |
| rs2072128545 | 1199 | H>N | No | gnomAD | |
| COSM1380748 | 1200 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs143575170 | 1201 | D>E | No |
ESP ExAC gnomAD |
|
| rs1367226499 | 1201 | D>G | No |
TOPMed gnomAD |
|
|
rs2072128281 TCGA novel |
1203 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs2072128318 | 1203 | M>L | No | gnomAD | |
| TCGA novel | 1203 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1048418522 | 1204 | A>T | No | Ensembl | |
| rs2072128188 | 1205 | E>K | No |
TOPMed gnomAD |
|
| rs1480470715 | 1207 | G>A | No |
TOPMed gnomAD |
|
|
RCV002250091 rs571189404 |
1207 | G>R | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs2072127843 | 1208 | E>D | No | gnomAD | |
| rs2072127941 | 1208 | E>K | No | Ensembl | |
| rs1377164657 | 1209 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs772183801 | 1210 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs772183801 | 1210 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs2072127676 | 1212 | N>H | No | TOPMed | |
| COSM75524 | 1214 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1215 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072127552 | 1215 | R>Q | No | Ensembl | |
|
COSM1380747 rs1403404409 |
1215 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
| TCGA novel | 1216 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1597399150 | 1216 | V>G | No | Ensembl | |
| rs185688883 | 1217 | K>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2072127382 | 1218 | Q>H | No | Ensembl | |
| COSM975196 | 1221 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs866990183 | 1222 | K>Q | No | Ensembl | |
| rs912354941 | 1222 | K>R | No |
TOPMed gnomAD |
|
| rs202177216 | 1224 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1470274648 | 1225 | S>R | No | gnomAD | |
| rs2072127096 | 1225 | S>T | No | Ensembl | |
| rs368839272 | 1228 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1283060152 | 1230 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1032490997 | 1231 | T>A | No | Ensembl | |
| rs2142174526 | 1231 | T>I | No | Ensembl | |
| rs1396008788 | 1232 | D>G | No |
TOPMed gnomAD |
|
| rs2142174519 | 1232 | D>N | No | Ensembl | |
| rs866491586 | 1233 | D>N | No | Ensembl | |
| rs779913726 | 1234 | L>F | No |
ExAC gnomAD |
|
| COSM6145655 | 1234 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072126366 | 1235 | S>T | No | gnomAD | |
| rs1374283170 | 1237 | N>I | No | gnomAD | |
| rs2072126147 | 1238 | A>G | No | TOPMed | |
|
COSM1380746 rs2072126196 |
1238 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| COSM4063800 | 1240 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754138767 | 1244 | A>P | No |
ExAC gnomAD |
|
| rs966771807 | 1245 | K>Q | No |
TOPMed gnomAD |
|
| rs1461072775 | 1245 | K>T | No | gnomAD | |
| rs932511304 | 1250 | K>E | No |
TOPMed gnomAD |
|
|
COSM705182 rs772023579 |
1250 | K>N | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated ExAC gnomAD NCI-TCGA Cosmic |
| rs201773508 | 1250 | K>R | No | 1000Genomes | |
| rs2072116476 | 1251 | M>I | No |
TOPMed gnomAD |
|
| TCGA novel | 1252 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1253112363 | 1253 | R>H | No | Ensembl | |
| rs537351462 | 1254 | S>F | No |
1000Genomes ExAC gnomAD |
|
| rs756830106 | 1254 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs756830106 | 1254 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs777344448 | 1255 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs756473788 | 1257 | D>N | No |
ExAC gnomAD |
|
| COSM3818782 | 1257 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072116034 | 1258 | Q>L | No | TOPMed | |
| rs1810132411 | 1259 | V>A | No | TOPMed | |
| rs2142173718 | 1259 | V>L | No | Ensembl | |
| rs575944229 | 1260 | S>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2072115872 | 1261 | E>D | No | Ensembl | |
|
rs1063926 VAR_030208 |
1261 | E>G | No |
UniProt Ensembl dbSNP |
|
| rs140482097 | 1262 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs146793402 | 1263 | K>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1421931499 | 1267 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs2072115603 | 1268 | E>G | No | TOPMed | |
| rs1265844506 | 1268 | E>K | No | gnomAD | |
| rs1265844506 | 1268 | E>Q | No | gnomAD | |
| rs2072115603 | 1268 | E>V | No | TOPMed | |
| COSM4831525 | 1269 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1171937682 | 1269 | Q>R | No | TOPMed | |
| rs1302473205 | 1270 | Q>K | No |
TOPMed gnomAD |
|
|
COSM71748 rs536291125 |
1271 | R>Q | ovary [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs761488980 COSM1563422 |
1271 | R>W | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| TCGA novel | 1272 | L>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072115436 | 1273 | I>F | No | Ensembl | |
| rs913641047 | 1274 | N>D | No | Ensembl | |
| rs184089245 | 1274 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372019710 | 1274 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2142173632 | 1275 | D>Y | No | Ensembl | |
| rs770410493 | 1276 | L>H | No |
ExAC gnomAD |
|
| rs774333066 | 1276 | L>I | No |
ExAC gnomAD |
|
| rs1348715591 | 1278 | A>E | No | gnomAD | |
| COSM6145656 | 1280 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM6145657 | 1281 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1231836083 | 1281 | A>T | No | gnomAD | |
| rs1450573502 | 1281 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
rs748488880 COSM180551 |
1282 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs748488880 | 1282 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs375133768 | 1282 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1268282594 | 1287 | A>G | No | gnomAD | |
| rs1597398802 | 1287 | A>T | No | Ensembl | |
|
rs1268282594 COSM1380744 |
1287 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs2072105062 | 1288 | G>D | No | Ensembl | |
| rs2072105029 | 1289 | E>* | No | Ensembl | |
| rs748783415 | 1292 | R>P | No |
ExAC TOPMed gnomAD |
|
|
rs748783415 COSM1380743 |
1292 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs892269115 | 1294 | L>S | No |
TOPMed gnomAD |
|
| rs1345117942 | 1295 | D>G | No | gnomAD | |
| rs772611607 | 1296 | E>* | No |
ExAC gnomAD |
|
| rs1063927 | 1297 | K>Q | No |
ExAC gnomAD |
|
| rs1337119789 | 1298 | D>G | No | gnomAD | |
| rs1290639984 | 1299 | A>T | No | gnomAD | |
| rs2142172793 | 1300 | L>F | No | Ensembl | |
| rs201768657 | 1301 | V>A | No | TOPMed | |
| COSM3513645 | 1301 | V>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs747753372 | 1301 | V>I | No |
ExAC gnomAD |
|
| rs781047786 | 1302 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs373767969 | 1303 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1398231586 | 1304 | L>F | No |
TOPMed gnomAD |
|
| rs1398231586 | 1304 | L>V | No |
TOPMed gnomAD |
|
| rs747419064 | 1306 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs780667343 | 1307 | S>G | No |
ExAC gnomAD |
|
| rs758846469 | 1307 | S>I | No |
ExAC gnomAD |
|
| rs201161796 | 1307 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1159709746 | 1309 | Q>E | No | gnomAD | |
| rs1159709746 | 1309 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs866739385 | 1310 | A>E | No | Ensembl | |
| TCGA novel | 1310 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1310 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1045155798 | 1313 | Q>L | No |
TOPMed gnomAD |
|
| rs1045155798 | 1313 | Q>R | No |
TOPMed gnomAD |
|
| rs1378429212 | 1314 | Q>* | No |
TOPMed gnomAD |
|
| rs778859716 | 1315 | I>F | No |
ExAC gnomAD |
|
| rs757421757 | 1315 | I>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1316 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764343697 | 1317 | E>G | No | ExAC | |
| rs1164868270 | 1317 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1352458408 | 1320 | H>R | No | gnomAD | |
| rs759292164 | 1320 | H>Y | No |
ExAC gnomAD |
|
| rs1268351167 | 1321 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs920208145 | 1322 | L>V | No |
TOPMed gnomAD |
|
| rs751379339 | 1323 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1229464291 | 1326 | T>I | No | gnomAD | |
| rs2072103777 | 1327 | K>E | No | gnomAD | |
| rs753954315 | 1330 | N>D | No |
ExAC gnomAD |
|
| rs143643784 | 1331 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
|
rs751289543 COSM4063799 |
1331 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1405725101 | 1333 | A>V | No | gnomAD | |
|
COSM394659 rs140081412 |
1334 | H>Q | lung [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs142232788 | 1335 | A>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1451189663 | 1336 | L>P | No | gnomAD | |
| rs1447337519 | 1337 | Q>L | No | gnomAD | |
| rs1254479049 | 1338 | S>P | No | gnomAD | |
| rs1752545351 | 1339 | S>C | No | TOPMed | |
| rs201987005 | 1339 | S>P | No |
ExAC gnomAD |
|
| rs538081232 | 1340 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776303430 | 1340 | R>H | No |
ExAC gnomAD |
|
| rs538081232 | 1340 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs768361993 | 1341 | H>R | No |
ExAC gnomAD |
|
| rs2072101756 | 1343 | C>Y | No | gnomAD | |
|
COSM3421271 rs775346508 |
1344 | D>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs201407686 | 1347 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs548884643 | 1347 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1348 | E>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs148582560 | 1349 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1330312530 | 1349 | Q>R | No | gnomAD | |
| rs772297289 | 1350 | Y>C | No | TOPMed | |
| rs1210774754 | 1352 | E>A | No |
TOPMed gnomAD |
|
| rs71358299 | 1353 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs756255780 | 1354 | Q>E | No | ExAC | |
| rs756255780 | 1354 | Q>K | No | ExAC | |
| rs376632174 | 1354 | Q>R | No |
TOPMed gnomAD |
|
| rs144823011 | 1357 | K>R | No |
ESP TOPMed gnomAD |
|
| rs2072100903 | 1359 | E>K | No | TOPMed | |
| rs2072100802 | 1361 | Q>* | No | Ensembl | |
| rs1191430194 | 1362 | R>T | No | gnomAD | |
| rs1246525590 | 1363 | A>G | No |
TOPMed gnomAD |
|
| rs781524378 | 1363 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1246525590 | 1363 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs750262993 | 1365 | S>C | No |
ExAC TOPMed gnomAD |
|
| COSM6145659 | 1365 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765035903 | 1366 | K>E | No |
ExAC gnomAD |
|
| rs757269825 | 1366 | K>N | No |
ExAC gnomAD |
|
| rs2072100328 | 1366 | K>R | No | Ensembl | |
| rs1340138986 | 1367 | A>D | No | gnomAD | |
| rs940248214 | 1368 | N>I | No | Ensembl | |
| TCGA novel | 1370 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763689601 | 1370 | E>K | No |
ExAC gnomAD |
|
| rs2072100058 | 1371 | V>A | No | TOPMed | |
| rs1344529001 | 1371 | V>I | No | gnomAD | |
| rs1376516976 | 1372 | A>T | No | gnomAD | |
| rs1272910869 | 1372 | A>V | No | gnomAD | |
| rs908787635 | 1374 | W>C | No | TOPMed | |
| rs774977418 | 1375 | R>G | No |
ExAC gnomAD |
|
| COSM3513643 | 1375 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1359128255 | 1377 | K>N | No |
TOPMed gnomAD |
|
| rs200411635 | 1378 | Y>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs547440233 | 1378 | Y>H | No |
1000Genomes ExAC gnomAD |
|
| rs547440233 | 1378 | Y>N | No |
1000Genomes ExAC gnomAD |
|
|
rs749792075 COSM3513642 |
1379 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs749792075 | 1379 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs565417467 | 1380 | T>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs565417467 | 1380 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748266367 | 1382 | A>V | No |
ExAC gnomAD |
|
| rs1276502664 | 1384 | Q>* | No | TOPMed | |
| rs556963319 | 1384 | Q>R | No |
TOPMed gnomAD |
|
| rs745591903 | 1385 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs147143044 | 1385 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| COSM6080176 | 1385 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072099357 | 1386 | T>A | No | TOPMed | |
| rs2072099331 | 1386 | T>I | No | Ensembl | |
| rs2142172329 | 1387 | E>Q | No | Ensembl | |
| rs968454745 | 1388 | E>K | No |
TOPMed gnomAD |
|
| rs1022281228 | 1388 | E>V | No |
TOPMed gnomAD |
|
| rs1597398330 | 1389 | L>Q | No | Ensembl | |
| rs764174268 | 1390 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs764174268 | 1390 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs988068417 | 1391 | E>K | No |
TOPMed gnomAD |
|
| COSM3513641 | 1393 | K>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs143515663 | 1393 | K>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs763700322 | 1394 | K>E | No | Ensembl | |
| rs2072085548 | 1394 | K>R | No | Ensembl | |
| rs941923100 | 1395 | K>N | No | TOPMed | |
| rs1597397851 | 1395 | K>T | No | Ensembl | |
| rs749129059 | 1397 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM1258480 rs924415460 |
1399 | R>C | Variant assessed as Somatic; MODERATE impact. oesophagus [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
| rs368200953 | 1399 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072085254 | 1403 | A>G | No | TOPMed | |
| TCGA novel | 1405 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1567682404 | 1406 | H>L | No | Ensembl | |
| rs780750095 | 1406 | H>Q | No |
ExAC gnomAD |
|
| rs754505775 | 1407 | V>A | No |
ExAC gnomAD |
|
| rs983269198 | 1408 | E>K | No | gnomAD | |
| COSM975193 | 1409 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2142171059 | 1409 | A>V | No | Ensembl | |
|
COSM1216253 rs1320361315 |
1410 | V>A | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs536918310 | 1411 | N>S | No | gnomAD | |
| rs750806957 | 1412 | A>D | No |
ExAC TOPMed gnomAD |
|
|
COSM1563423 rs200412862 |
1412 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs750806957 RCV000906090 |
1412 | A>V | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| COSM3513640 | 1416 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs2142171029 COSM3513639 |
1417 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| COSM6145660 | 1418 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs150344258 COSM3402578 |
1420 | T>M | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1473287677 | 1420 | T>S | No | Ensembl | |
| rs2072084735 | 1421 | K>N | No |
TOPMed gnomAD |
|
|
rs768767249 COSM2924724 |
1423 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs777070777 | 1423 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs974863672 | 1425 | Q>* | No | Ensembl | |
| rs2072084648 | 1426 | N>D | No | Ensembl | |
| rs760715661 | 1426 | N>S | No |
ExAC gnomAD |
|
| rs772447154 | 1428 | V>A | No |
ExAC gnomAD |
|
| rs775820934 | 1428 | V>I | No |
ExAC gnomAD |
|
| rs2072084357 | 1430 | D>G | No | TOPMed | |
| rs777715204 | 1430 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1021682488 | 1432 | M>T | No |
TOPMed gnomAD |
|
| COSM2924723 | 1433 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1567682339 | 1434 | D>Y | No | Ensembl | |
| rs141278410 | 1435 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072084196 | 1436 | E>K | No | TOPMed | |
| rs1597397796 | 1437 | R>G | No | Ensembl | |
| rs2072084130 | 1437 | R>M | No | Ensembl | |
| rs1294451994 | 1437 | R>S | No | gnomAD | |
| rs781044002 | 1438 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs960021077 | 1439 | N>H | No |
TOPMed gnomAD |
|
| rs754482819 | 1440 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM4893481 | 1442 | C>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072083896 | 1444 | A>T | No | gnomAD | |
| rs779629902 | 1446 | D>H | No |
ExAC TOPMed gnomAD |
|
|
rs779629902 COSM3513637 |
1446 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1447 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1435075312 | 1450 | R>W | No | gnomAD | |
| rs2072083589 | 1451 | N>I | No | Ensembl | |
| rs551879041 | 1452 | F>S | No | Ensembl | |
| COSM1479204 | 1453 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072082927 | 1454 | K>E | No | Ensembl | |
| rs1370047465 | 1455 | V>I | No | gnomAD | |
| rs1372272655 | 1457 | S>P | No | gnomAD | |
| rs1567682279 | 1458 | E>A | No | Ensembl | |
| rs1166186246 | 1459 | W>* | No | gnomAD | |
| rs1474422672 | 1459 | W>R | No | Ensembl | |
|
RCV000881615 CA8387291 RCV000253582 rs112910113 |
1460 | K>R | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2142170790 | 1463 | Y>H | No | Ensembl | |
| rs1479337413 | 1464 | E>D | No |
TOPMed gnomAD |
|
| rs759834838 | 1464 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs759834838 | 1464 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1413830867 | 1466 | T>I | No | gnomAD | |
| rs774775779 | 1467 | Q>R | No |
ExAC gnomAD |
|
| TCGA novel | 1468 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072081547 | 1469 | E>D | No | Ensembl | |
| rs144853263 | 1469 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072081650 | 1469 | E>K | No | gnomAD | |
| rs1201657302 | 1470 | L>F | No | gnomAD | |
| rs1483407920 | 1470 | L>H | No |
TOPMed gnomAD |
|
| TCGA novel | 1471 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751090587 | 1471 | E>Q | No |
TOPMed gnomAD |
|
| rs199592324 | 1472 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs776594201 | 1472 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs867618980 | 1473 | S>F | No | Ensembl | |
| rs1321869015 | 1474 | Q>R | No | gnomAD | |
| rs867112413 | 1475 | K>N | No | Ensembl | |
|
COSM1493790 rs1319472947 |
1476 | E>G | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs768497869 | 1478 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs2072081117 | 1479 | S>T | No | Ensembl | |
| rs1597397700 | 1480 | L>V | No | Ensembl | |
| COSM4063798 | 1481 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1326987976 | 1482 | T>I | No | gnomAD | |
| TCGA novel | 1482 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1397842983 | 1485 | F>L | No | gnomAD | |
| rs2072080988 | 1488 | K>N | No | TOPMed | |
|
rs757966246 COSM3513636 |
1489 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1468388124 | 1490 | V>A | No | gnomAD | |
| rs1336339178 | 1490 | V>I | No | gnomAD | |
|
rs1060499730 CA16609480 RCV000596881 |
1491 | Y>missing | No |
ClinGen ClinVar dbSNP |
|
| rs376996294 | 1491 | Y>C | No |
1000Genomes ESP ExAC gnomAD |
|
| rs2142170677 | 1492 | E>G | No | Ensembl | |
| rs757608378 | 1493 | E>G | No |
ExAC gnomAD |
|
| TCGA novel | 1493 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764606216 | 1494 | S>C | No |
ExAC gnomAD |
|
| rs764606216 | 1494 | S>Y | No |
ExAC gnomAD |
|
| rs756584687 | 1495 | L>P | No |
ExAC gnomAD |
|
| rs756584687 | 1495 | L>R | No |
ExAC gnomAD |
|
| rs1428994626 | 1496 | D>G | No |
TOPMed gnomAD |
|
| rs557729639 | 1497 | Q>R | No |
1000Genomes ExAC gnomAD |
|
| rs1201409140 | 1498 | L>H | No |
TOPMed gnomAD |
|
|
COSM3513635 rs370499944 |
1499 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs73977155 | 1500 | T>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs761617178 | 1501 | L>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1502 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1247248833 | 1502 | R>K | No |
TOPMed gnomAD |
|
| rs1140936 | 1504 | E>A | No |
ExAC gnomAD |
|
| rs1140936 | 1504 | E>G | No |
ExAC gnomAD |
|
| rs1392348379 | 1504 | E>K | No | gnomAD | |
| rs768568873 | 1507 | N>S | No |
ExAC gnomAD |
|
| rs768568873 | 1507 | N>T | No |
ExAC gnomAD |
|
| rs1453651895 | 1508 | L>S | No |
TOPMed gnomAD |
|
| rs202040130 | 1510 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| COSM975191 | 1510 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072079186 | 1511 | E>D | No | Ensembl | |
| rs1214941044 | 1511 | E>G | No | gnomAD | |
| rs1597397604 | 1512 | I>T | No | Ensembl | |
|
CA497838201 rs1459072387 RCV000500131 |
1513 | S>missing | No |
ClinGen ClinVar dbSNP |
|
| rs2072079102 | 1513 | S>C | No | TOPMed | |
| rs2072079102 | 1513 | S>F | No | TOPMed | |
|
rs763087671 COSM211830 |
1514 | D>E | breast [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs766563318 | 1514 | D>Y | No |
ExAC gnomAD |
|
|
RCV001354725 rs34564342 |
1515 | L>I | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1364860639 | 1516 | T>I | No | gnomAD | |
| rs1161192269 | 1517 | E>D | No | gnomAD | |
| rs1431098476 | 1518 | Q>H | No | gnomAD | |
| rs2072078902 | 1518 | Q>K | No |
1000Genomes TOPMed |
|
| rs2072078871 | 1518 | Q>R | No | Ensembl | |
| rs145021815 | 1519 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1390183613 | 1519 | I>V | No | gnomAD | |
| rs2142170440 | 1520 | A>V | No | Ensembl | |
| rs1461413222 | 1521 | E>G | No | TOPMed | |
| rs2072078729 | 1521 | E>K | No | Ensembl | |
| rs763779942 | 1523 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1324442337 | 1523 | G>R | No | Ensembl | |
|
rs775373801 COSM472266 |
1525 | Q>* | Variant assessed as Somatic; HIGH impact. kidney [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed |
| rs775373801 | 1525 | Q>K | No |
ExAC TOPMed |
|
| rs772027441 | 1526 | I>N | No |
ExAC gnomAD |
|
| rs1220822681 | 1529 | L>W | No | gnomAD | |
| rs773740111 | 1531 | K>* | No |
ExAC gnomAD |
|
| rs2072078376 | 1531 | K>T | No | Ensembl | |
| COSM3969793 | 1535 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1306298843 | 1537 | E>K | No | TOPMed | |
| rs770542655 | 1538 | Q>E | No |
ExAC gnomAD |
|
| rs2072078211 | 1538 | Q>H | No | gnomAD | |
| rs770542655 | 1538 | Q>K | No |
ExAC gnomAD |
|
| rs1337657555 | 1539 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 1541 | C>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs868298868 COSM5385632 |
1542 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs2072078135 | 1544 | Q>* | No | TOPMed | |
| rs748880308 | 1545 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1273626653 | 1546 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| COSM6080178 | 1548 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1436914559 | 1549 | E>K | No | gnomAD | |
| rs1344875858 | 1550 | A>E | No |
TOPMed gnomAD |
|
|
RCV001567092 rs1344875858 |
1550 | A>G | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1344875858 | 1550 | A>V | No |
TOPMed gnomAD |
|
| rs1301220270 | 1551 | E>K | No | gnomAD | |
|
COSM1479203 rs762568851 |
1552 | A>P | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs762568851 | 1552 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs2072070516 | 1552 | A>V | No | Ensembl | |
| rs772703670 | 1553 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs772703670 | 1553 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs769596464 | 1556 | H>D | No |
ExAC TOPMed gnomAD |
|
| rs769596464 | 1556 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1474354062 | 1557 | E>Q | No | TOPMed | |
| rs2072070322 | 1558 | E>* | No | TOPMed | |
| rs2072070289 | 1558 | E>D | No | TOPMed | |
| rs1567681820 | 1560 | K>R | No | Ensembl | |
| rs781691482 | 1561 | I>S | No |
ExAC TOPMed gnomAD |
|
| rs781691482 | 1561 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1395158149 | 1562 | L>P | No | TOPMed | |
|
COSM975190 rs1433759882 |
1563 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs544591533 | 1563 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM1479202 | 1564 | I>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1176755745 | 1565 | Q>H | No |
TOPMed gnomAD |
|
| rs2142169682 | 1567 | E>G | No | Ensembl | |
| rs780656237 | 1568 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs758377642 | 1569 | N>T | No |
ExAC gnomAD |
|
| COSM3795258 | 1569 | N>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1461015140 | 1570 | Q>P | No | gnomAD | |
| TCGA novel | 1571 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1261076662 | 1571 | V>I | No | gnomAD | |
| rs61730805 | 1575 | V>G | No | gnomAD | |
| TCGA novel | 1579 | I>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs181695343 | 1580 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1380744160 | 1580 | A>V | No | gnomAD | |
| rs1451509674 | 1582 | K>N | No | gnomAD | |
| rs1380089220 | 1586 | I>T | No | gnomAD | |
| rs2072069518 | 1587 | D>E | No | Ensembl | |
| rs1567681771 | 1590 | K>M | No | Ensembl | |
| rs1459150106 | 1590 | K>N | No | gnomAD | |
| rs751421547 | 1593 | H>L | No |
ExAC gnomAD |
|
| rs2142169601 | 1593 | H>N | No | Ensembl | |
| rs2072069352 | 1594 | T>N | No | Ensembl | |
| rs1420526673 | 1595 | R>G | No | gnomAD | |
|
RCV000971236 rs145711576 |
1595 | R>T | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs61730807 | 1596 | V>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1006892268 | 1597 | V>A | No |
TOPMed gnomAD |
|
| rs369298245 | 1597 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs369298245 | 1597 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072069092 | 1598 | E>Q | No | TOPMed | |
| rs1486081243 | 1601 | Q>* | No |
TOPMed gnomAD |
|
| rs1486081243 | 1601 | Q>K | No |
TOPMed gnomAD |
|
| rs775820322 | 1603 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1159573863 | 1606 | A>V | No | TOPMed | |
| COSM975189 | 1607 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1384657399 | 1608 | I>M | No |
TOPMed gnomAD |
|
| rs1456156935 | 1609 | R>S | No |
TOPMed gnomAD |
|
| rs2072068764 | 1610 | S>G | No | gnomAD | |
|
rs144447823 COSM107360 |
1611 | R>K | large_intestine skin [Cosmic] | No |
cosmic curated Ensembl |
| rs1291926389 | 1611 | R>S | No | gnomAD | |
| COSM6080181 | 1613 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1051333342 | 1614 | A>G | No |
TOPMed gnomAD |
|
| rs1051333342 | 1614 | A>V | No |
TOPMed gnomAD |
|
| rs2072068652 | 1615 | L>P | No | Ensembl | |
|
rs778961328 CA8387196 RCV000591499 |
1616 | R>* | No |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
| rs1356907051 | 1616 | R>K | No | gnomAD | |
| rs757387520 | 1617 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs757387520 | 1617 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs200572244 | 1618 | K>E | No |
1000Genomes ExAC gnomAD |
|
| rs1363118111 | 1618 | K>N | No | gnomAD | |
| rs1567681727 | 1618 | K>R | No | Ensembl | |
| rs201775710 | 1620 | K>E | No | 1000Genomes | |
| rs774191268 | 1621 | M>IIKV* | No | ExAC | |
| rs778011337 | 1622 | E>G | No |
ExAC gnomAD |
|
| rs1231849439 | 1624 | D>A | No |
TOPMed gnomAD |
|
| rs1017942863 | 1624 | D>N | No | gnomAD | |
| COSM1380741 | 1626 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs202123184 | 1626 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1626 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM705184 | 1627 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072068096 | 1628 | M>I | No |
TOPMed gnomAD |
|
| rs151301625 | 1628 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1278781469 | 1629 | E>Q | No | TOPMed | |
| rs2072068064 | 1630 | I>M | No | Ensembl | |
|
TCGA novel rs2072068039 |
1631 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| COSM705185 | 1631 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1326789382 | 1631 | Q>R | No |
TOPMed gnomAD |
|
| rs1439143145 | 1632 | L>V | No | gnomAD | |
| rs1567681701 | 1633 | N>I | No | Ensembl | |
| rs1274948091 | 1634 | H>R | No | gnomAD | |
| COSM975188 | 1635 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs750448710 | 1635 | A>V | No |
ExAC gnomAD |
|
| rs764824437 | 1636 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs866728917 | 1636 | N>S | No |
TOPMed gnomAD |
|
| rs866728917 | 1636 | N>T | No |
TOPMed gnomAD |
|
| rs544145292 | 1637 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs544145292 | 1637 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs544145292 | 1637 | R>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1641 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763838601 | 1642 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1356659510 | 1644 | R>K | No | gnomAD | |
| rs368308961 | 1646 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| COSM1135866 | 1646 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072067493 | 1646 | Y>H | No | gnomAD | |
| rs2072067404 | 1648 | N>K | No | TOPMed | |
| rs775934262 | 1649 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs375724202 | 1651 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2072067283 | 1653 | L>R | No | Ensembl | |
| COSM4893438 | 1654 | K>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1655 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764767647 | 1660 | L>P | No | Ensembl | |
| rs1175404788 | 1663 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| TCGA novel | 1664 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs367601078 | 1665 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072061187 | 1666 | G>A | No | gnomAD | |
| rs1247748406 | 1667 | Q>P | No | gnomAD | |
| rs1201857798 | 1668 | E>K | No | gnomAD | |
| rs2072061021 | 1669 | D>Y | No | TOPMed | |
| rs1461467864 | 1670 | L>F | No |
TOPMed gnomAD |
|
| rs1461467864 | 1670 | L>I | No |
TOPMed gnomAD |
|
| rs2072060914 | 1671 | K>R | No | TOPMed | |
| rs1203414443 | 1672 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2072060785 | 1673 | Q>E | No |
TOPMed gnomAD |
|
| rs2072060750 | 1673 | Q>P | No | Ensembl | |
|
rs1322113245 COSM1493791 |
1675 | A>V | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1024111495 | 1676 | I>T | No | gnomAD | |
| rs557187036 | 1677 | V>A | No |
1000Genomes ExAC gnomAD |
|
| rs2072060399 | 1678 | E>D | No | TOPMed | |
|
rs370435018 COSM975187 |
1679 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs202001342 | 1679 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs370435018 | 1679 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072060177 | 1680 | R>G | No | gnomAD | |
| rs1034056044 | 1684 | L>P | No |
TOPMed gnomAD |
|
| rs1034056044 | 1684 | L>Q | No |
TOPMed gnomAD |
|
| rs1034056044 | 1684 | L>R | No |
TOPMed gnomAD |
|
| rs2072059997 | 1685 | Q>R | No | gnomAD | |
| rs374032538 | 1687 | E>K | No |
ESP ExAC gnomAD |
|
| rs1049319962 | 1689 | E>G | No | Ensembl | |
| rs755630877 | 1689 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs755630877 | 1689 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1484891697 | 1691 | L>M | No |
TOPMed gnomAD |
|
| rs752365419 | 1691 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs147139573 | 1692 | W>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1203255970 | 1695 | L>P | No |
TOPMed gnomAD |
|
| rs1460268562 | 1695 | L>V | No |
TOPMed gnomAD |
|
| rs2072059450 | 1696 | E>K | No | Ensembl | |
| COSM6080182 | 1699 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072059268 | 1699 | E>D | No | TOPMed | |
| TCGA novel | 1700 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072059210 | 1701 | S>R | No | TOPMed | |
|
COSM1709823 rs2142168714 |
1702 | R>K | skin [Cosmic] | No |
cosmic curated Ensembl |
| rs140525529 | 1702 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs143448404 | 1705 | A>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA205926 rs143448404 RCV000192831 |
1705 | A>T | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2072058970 | 1706 | E>G | No | Ensembl | |
| rs776869211 | 1706 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs776869211 | 1706 | E>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1707 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM705187 | 1707 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1450660738 | 1707 | Q>R | No |
TOPMed gnomAD |
|
| rs745600958 | 1708 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs745600958 | 1708 | E>Q | No |
ExAC TOPMed gnomAD |
|
|
rs148425060 COSM435981 |
1709 | L>F | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs749216619 | 1712 | A>G | No |
ExAC gnomAD |
|
| COSM705188 | 1712 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs144477258 | 1714 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs553069572 | 1714 | E>K | No |
TOPMed gnomAD |
|
| rs376736272 | 1715 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM3402577 rs780844078 |
1715 | R>H | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed |
| rs1450899315 | 1716 | V>A | No |
TOPMed gnomAD |
|
| TCGA novel | 1716 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754634113 | 1716 | V>I | No |
ExAC gnomAD |
|
|
rs766719917 COSM1380739 |
1719 | L>I | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA |
| rs1194317809 | 1721 | T>A | No | gnomAD | |
| rs2072058382 | 1722 | Q>H | No | TOPMed | |
| COSM1380738 | 1722 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1284920104 | 1724 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| TCGA novel | 1726 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750772829 | 1730 | K>* | No |
ExAC TOPMed gnomAD |
|
| rs754435722 | 1735 | N>S | No |
ExAC gnomAD |
|
| rs33969260 | 1736 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1459255088 | 1737 | V>A | No | TOPMed | |
| rs775814737 | 1737 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs767877486 | 1739 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2072046083 | 1739 | Q>P | No | Ensembl | |
| rs769699528 | 1740 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs769699528 | 1740 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2072045966 | 1741 | Q>R | No | TOPMed | |
| rs768120382 | 1742 | S>R | No |
ExAC gnomAD |
|
| rs374898030 | 1742 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs746549585 | 1744 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs779629920 | 1746 | E>* | No |
ExAC gnomAD |
|
| rs771633535 | 1746 | E>D | No |
ExAC gnomAD |
|
| rs2072045758 | 1747 | V>I | No | TOPMed | |
| rs2072045758 | 1747 | V>L | No | TOPMed | |
| rs746175156 | 1750 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs746175156 | 1750 | E>Q | No |
ExAC TOPMed gnomAD |
|
|
rs779138246 COSM180545 |
1752 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs779138246 | 1752 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs34433607 | 1752 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs34433607 | 1752 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2072045550 | 1753 | N>K | No | Ensembl | |
| rs2072045521 | 1754 | A>T | No |
TOPMed gnomAD |
|
| TCGA novel | 1754 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1756 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1756 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1756 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1396942164 | 1757 | K>* | No | Ensembl | |
| rs2072045378 | 1758 | A>G | No | TOPMed | |
| rs1281134325 | 1760 | K>E | No |
TOPMed gnomAD |
|
| rs1439631679 | 1760 | K>R | No | gnomAD | |
| rs1336565117 | 1761 | A>D | No | Ensembl | |
| rs563469677 | 1761 | A>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs563469677 | 1761 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3513627 | 1761 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2072045245 | 1762 | I>T | No | Ensembl | |
| rs756213751 | 1763 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs756213751 | 1763 | T>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1764 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2142167456 | 1764 | D>Y | No | Ensembl | |
| rs1158563369 | 1765 | A>D | No | gnomAD | |
| rs749692404 | 1765 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1158563369 | 1765 | A>V | No | gnomAD | |
| rs771654542 | 1766 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs2072043796 | 1766 | A>V | No | TOPMed | |
| rs1387104661 | 1767 | M>T | No | gnomAD | |
| rs2072043741 | 1767 | M>V | No | TOPMed | |
| rs267604699 | 1768 | M>I | No | Ensembl | |
| rs2072043672 | 1768 | M>T | No |
TOPMed gnomAD |
|
| COSM1709822 | 1771 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs199803469 | 1771 | E>Q | No |
1000Genomes TOPMed gnomAD |
|
| rs371184733 | 1773 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001093233 rs753326378 COSM3889211 |
1775 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs1241447824 | 1776 | Q>E | No | gnomAD | |
| rs577510735 | 1777 | D>G | No |
1000Genomes ExAC gnomAD |
|
| rs1244845907 | 1778 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1319532143 | 1778 | T>I | No | gnomAD | |
| rs1319532143 | 1778 | T>N | No | gnomAD | |
| rs751704006 | 1779 | S>G | No |
ExAC gnomAD |
|
| TCGA novel | 1779 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs200906285 COSM3421270 |
1780 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1597396580 | 1781 | H>P | No | Ensembl | |
| rs1396330990 | 1781 | H>Y | No |
TOPMed gnomAD |
|
| rs1288810481 | 1782 | L>R | No | gnomAD | |
| rs1398649581 | 1783 | E>K | No | gnomAD | |
| rs138165573 | 1784 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs141215006 RCV000658770 |
1784 | R>W | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2142167247 | 1785 | M>T | No | Ensembl | |
| rs1597396561 | 1788 | N>T | No | Ensembl | |
| rs1597396558 | 1789 | L>R | No | Ensembl | |
| TCGA novel | 1790 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1437236803 | 1790 | E>G | No | gnomAD | |
| rs1287040846 | 1790 | E>K | No | gnomAD | |
|
rs144962215 COSM3733901 |
1792 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1793 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072042203 | 1793 | V>G | No | Ensembl | |
| rs1597396541 | 1793 | V>M | No | Ensembl | |
| rs2072042156 | 1794 | K>E | No | Ensembl | |
| rs1597396536 | 1795 | D>A | No | Ensembl | |
| rs2072042073 | 1795 | D>N | No | Ensembl | |
| rs1369109003 | 1796 | L>P | No | Ensembl | |
| rs770630534 | 1796 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1472481825 | 1797 | Q>K | No |
TOPMed gnomAD |
|
| rs1221079892 | 1797 | Q>R | No | gnomAD | |
| rs1487340735 | 1798 | H>L | No | gnomAD | |
| rs749599374 | 1798 | H>Q | No |
ExAC gnomAD |
|
|
COSM559519 rs1204708839 |
1799 | R>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1204708839 | 1799 | R>G | No |
TOPMed gnomAD |
|
| rs773708880 | 1799 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1204708839 | 1799 | R>S | No |
TOPMed gnomAD |
|
| rs754623813 | 1800 | L>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1801 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1394332975 | 1802 | E>V | No | TOPMed | |
| COSM3513626 | 1803 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1404412785 | 1804 | E>D | No | gnomAD | |
| rs781745071 | 1804 | E>K | No |
ExAC gnomAD |
|
| rs1339393133 | 1805 | Q>H | No | Ensembl | |
| rs1313276726 | 1805 | Q>K | No |
TOPMed gnomAD |
|
| rs1300132926 | 1807 | A>E | No |
TOPMed gnomAD |
|
| rs1243043490 | 1807 | A>S | No |
TOPMed gnomAD |
|
| rs1300132926 | 1807 | A>V | No |
TOPMed gnomAD |
|
| rs1342415960 | 1809 | K>Q | No |
TOPMed gnomAD |
|
| rs1597396499 | 1810 | G>C | No |
TOPMed gnomAD |
|
| rs1597396499 | 1810 | G>S | No |
TOPMed gnomAD |
|
| rs62058111 | 1810 | G>V | No | Ensembl | |
| rs1168248353 | 1811 | G>A | No | gnomAD | |
| rs200167639 | 1813 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs190632125 | 1814 | Q>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3513624 | 1814 | Q>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752544677 | 1819 | E>K | No |
ExAC TOPMed gnomAD |
|
| COSM3513623 | 1820 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1480094123 | 1821 | R>K | No |
TOPMed gnomAD |
|
| rs143876651 | 1822 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs530187127 | 1823 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs202234534 | 1823 | R>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1823 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs113587727 | 1824 | E>G | No | Ensembl | |
| rs762594623 | 1824 | E>K | No |
ExAC gnomAD |
|
| rs772962350 | 1828 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs762206560 | 1829 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2072037087 | 1829 | V>I | No | Ensembl | |
| COSM3513622 | 1830 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1452443024 | 1831 | N>S | No | gnomAD | |
| rs72814800 | 1835 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA gnomAD |
| rs777055080 | 1835 | R>L | No |
ExAC TOPMed gnomAD |
|
| COSM6145662 | 1838 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs145809088 | 1838 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs772107409 | 1839 | A>T | No | ExAC | |
| rs745978736 | 1839 | A>V | No |
ExAC gnomAD |
|
| rs79577802 | 1840 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs79577802 | 1840 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs576121941 | 1840 | V>I | No |
1000Genomes ExAC gnomAD |
|
| rs576121941 | 1840 | V>L | No |
1000Genomes ExAC gnomAD |
|
| rs2072036498 | 1840 | V>Y | No | Ensembl | |
| rs562027482 | 1842 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs562027482 | 1842 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1242345525 | 1844 | R>L | No |
1000Genomes TOPMed gnomAD |
|
|
rs1242345525 COSM1380737 |
1844 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes NCI-TCGA TOPMed gnomAD |
|
COSM3402576 rs751486450 |
1844 | R>W | central_nervous_system [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1351330187 | 1846 | H>P | No |
TOPMed gnomAD |
|
| rs199753332 | 1846 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1053920 | 1847 | E>D | No | Ensembl | |
|
COSM215668 rs764912497 |
1848 | R>* | central_nervous_system [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs201648248 | 1849 | R>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM3513621 rs553671041 |
1849 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1597396345 | 1850 | V>G | No | Ensembl | |
| rs2072035756 | 1851 | K>N | No | Ensembl | |
| rs2072035733 | 1852 | E>* | No |
TOPMed gnomAD |
|
| rs2072035733 | 1852 | E>Q | No |
TOPMed gnomAD |
|
| rs1375094521 | 1855 | Y>* | No | gnomAD | |
| rs2072029597 | 1858 | E>K | No | TOPMed | |
| rs2072029597 | 1858 | E>Q | No | TOPMed | |
| rs1422113193 | 1859 | E>G | No | gnomAD | |
| rs1039866740 | 1859 | E>K | No |
TOPMed gnomAD |
|
| COSM1380736 | 1860 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs778549995 | 1860 | D>G | No |
ExAC gnomAD |
|
| rs1202153004 | 1860 | D>H | No | gnomAD | |
| rs778549995 | 1860 | D>V | No |
ExAC gnomAD |
|
| rs760517620 | 1861 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs1197069498 COSM975186 |
1861 | R>H | endometrium [Cosmic] | No |
cosmic curated gnomAD |
| rs2072029195 | 1865 | L>F | No | TOPMed | |
| rs763684063 | 1865 | L>H | No |
ExAC TOPMed gnomAD |
|
| rs1197778252 | 1866 | R>S | No | gnomAD | |
| rs761050897 | 1867 | L>M | No |
ExAC gnomAD |
|
| rs773251493 | 1868 | Q>* | No | Ensembl | |
| rs2072028935 | 1869 | D>H | No | Ensembl | |
| rs1223993210 | 1870 | L>V | No | gnomAD | |
| rs1343714566 | 1871 | V>A | No | gnomAD | |
|
CA16609479 rs1060499727 RCV000593052 |
1875 | Q>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs2072028556 | 1875 | Q>H | No | TOPMed | |
| rs1443990194 | 1875 | Q>R | No | gnomAD | |
| rs200825271 | 1876 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200825271 | 1876 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs762977268 | 1877 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs190701829 | 1877 | K>N | No |
1000Genomes ExAC gnomAD |
|
| rs1398494147 | 1877 | K>R | No | gnomAD | |
| rs1415511860 | 1878 | V>M | No | gnomAD | |
| TCGA novel | 1879 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769925803 | 1879 | K>I | No |
ExAC TOPMed gnomAD |
|
| rs769925803 | 1879 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs2072028166 | 1880 | S>* | No | TOPMed | |
| COSM3513620 | 1880 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs367910491 | 1881 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1471320473 | 1881 | Y>C | No | gnomAD | |
| TCGA novel | 1882 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM6145663 | 1883 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1244705458 | 1883 | R>K | No | gnomAD | |
| rs779032196 | 1885 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs771893150 | 1885 | A>V | No | Ensembl | |
| COSM705190 | 1886 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1275965939 | 1886 | E>K | No | gnomAD | |
| rs2072027659 | 1887 | E>A | No | TOPMed | |
| rs1224968997 | 1888 | A>S | No | gnomAD | |
| rs756750445 | 1888 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs1229257770 COSM3889210 |
1889 | E>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| TCGA novel | 1890 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1378831161 | 1890 | E>G | No | gnomAD | |
| rs2072011491 | 1891 | Q>K | No | TOPMed | |
| rs1282969866 | 1891 | Q>R | No | gnomAD | |
| rs267604697 | 1892 | S>F | No | Ensembl | |
| rs771959591 | 1893 | N>H | No |
ExAC gnomAD |
|
| rs2072011323 | 1894 | A>T | No | TOPMed | |
| rs368842417 | 1897 | S>P | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1899 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2142165056 | 1899 | F>V | No | Ensembl | |
| rs1165675056 | 1900 | R>C | No |
TOPMed gnomAD |
|
| rs770787118 | 1900 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs770787118 | 1900 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1903 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749314949 | 1904 | H>Y | No |
ExAC gnomAD |
|
| rs777135750 | 1905 | E>A | No |
ExAC gnomAD |
|
| rs1004389934 | 1907 | E>G | No | Ensembl | |
| rs1261906511 | 1908 | E>A | No | gnomAD | |
| COSM975185 | 1909 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs374807428 | 1910 | E>A | No |
ESP TOPMed gnomAD |
|
| COSM1380735 | 1910 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs755283265 | 1910 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs766839152 | 1912 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs751931786 | 1912 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2072010675 | 1915 | I>T | No | TOPMed | |
| rs1404868874 | 1916 | A>V | No |
TOPMed gnomAD |
|
| rs751000838 | 1919 | Q>* | No | ExAC | |
| rs2072010582 | 1919 | Q>H | No |
TOPMed gnomAD |
|
| rs1341667259 | 1921 | N>D | No | gnomAD | |
| rs1395882757 | 1923 | L>S | No |
TOPMed gnomAD |
|
| rs1398243359 | 1924 | R>* | No |
TOPMed gnomAD |
|
| rs1398243359 | 1924 | R>G | No |
TOPMed gnomAD |
|
| rs762026764 | 1924 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs762026764 COSM3513619 |
1924 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| COSM1324198 | 1925 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1567679851 | 1925 | V>M | No | Ensembl | |
| rs1457567748 | 1927 | S>R | No |
TOPMed gnomAD |
|
| rs374253045 | 1928 | R>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs374253045 | 1928 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1316167351 | 1928 | R>P | No |
TOPMed gnomAD |
|
| rs1316167351 | 1928 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1172249804 | 1930 | V>F | No |
TOPMed gnomAD |
|
|
RCV000733689 rs1346984851 |
1930 | V>G | No |
ClinVar dbSNP gnomAD |
|
| rs1172249804 | 1930 | V>I | No |
TOPMed gnomAD |
|
| rs2072010042 | 1932 | T>P | No | Ensembl | |
| rs934091317 | 1934 | I>V | No |
TOPMed gnomAD |
|
| COSM705191 | 1935 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372307102 | 1935 | S>N | No |
ESP ExAC gnomAD |
|
| rs770816590 | 1937 | E>K | No |
ExAC gnomAD |
2 associated diseases with P13535
[MIM: 608837]: Carney complex variant (CACOV)
Carney complex is a multiple neoplasia syndrome characterized by spotty skin pigmentation, cardiac and other myxomas, endocrine tumors, and psammomatous melanotic schwannomas. Familial cardiac myxomas are associated with spotty pigmentation of the skin and other phenotypes, including primary pigmented nodular adrenocortical dysplasia, extracardiac (frequently cutaneous) myxomas, schwannomas, and pituitary, thyroid, testicular, bone, ovarian, and breast tumors. Cardiac myxomas do not develop in all patients with the Carney complex, but affected patients have at least two features of the complex or one feature and a clinically significant family history. . Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 158300]: Arthrogryposis, distal, 7 (DA7)
A form of distal arthrogryposis, a disease characterized by congenital joint contractures that mainly involve two or more distal parts of the limbs, in the absence of a primary neurological or muscle disease. DA7 is characterized by an inability to open the mouth fully (trismus) and pseudocamptodactyly in which wrist dorsiflexion, but not volarflexion, produces involuntary flexion contracture of distal and proximal interphalangeal joints. Additional features include shortened hamstring muscles and short stature. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- Carney complex is a multiple neoplasia syndrome characterized by spotty skin pigmentation, cardiac and other myxomas, endocrine tumors, and psammomatous melanotic schwannomas. Familial cardiac myxomas are associated with spotty pigmentation of the skin and other phenotypes, including primary pigmented nodular adrenocortical dysplasia, extracardiac (frequently cutaneous) myxomas, schwannomas, and pituitary, thyroid, testicular, bone, ovarian, and breast tumors. Cardiac myxomas do not develop in all patients with the Carney complex, but affected patients have at least two features of the complex or one feature and a clinically significant family history. . Note=The disease is caused by variants affecting the gene represented in this entry.
- A form of distal arthrogryposis, a disease characterized by congenital joint contractures that mainly involve two or more distal parts of the limbs, in the absence of a primary neurological or muscle disease. DA7 is characterized by an inability to open the mouth fully (trismus) and pseudocamptodactyly in which wrist dorsiflexion, but not volarflexion, produces involuntary flexion contracture of distal and proximal interphalangeal joints. Additional features include shortened hamstring muscles and short stature. . Note=The disease is caused by variants affecting the gene represented in this entry.
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| muscle myosin complex | A filament of myosin found in a muscle cell of any type. |
| myosin filament | A supramolecular fiber containing myosin heavy chains, plus associated light chains and other proteins, in which the myosin heavy chains are arranged into a filament. |
| myosin II complex | A myosin complex containing two class II myosin heavy chains, two myosin essential light chains and two myosin regulatory light chains. Also known as classical myosin or conventional myosin, the myosin II class includes the major muscle myosin of vertebrate and invertebrate muscle, and is characterized by alpha-helical coiled coil tails that self assemble to form a variety of filament structures. |
| sarcomere | The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| microfilament motor activity | A motor activity that generates movement along a microfilament, driven by ATP hydrolysis. |
| myosin light chain binding | Binding to a light chain of a myosin complex. |
| myosin phosphatase activity | Catalysis of the reaction |
| structural constituent of muscle | The action of a molecule that contributes to the structural integrity of a muscle fiber. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| ATP metabolic process | The chemical reactions and pathways involving ATP, adenosine triphosphate, a universally important coenzyme and enzyme regulator. |
| muscle contraction | A process in which force is generated within muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis. |
| muscle filament sliding | The sliding of actin thin filaments and myosin thick filaments past each other in muscle contraction. This involves a process of interaction of myosin located on a thick filament with actin located on a thin filament. During this process ATP is split and forces are generated. |
| skeletal muscle contraction | A process in which force is generated within skeletal muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis. In the skeletal muscle, the muscle contraction takes advantage of an ordered sarcomeric structure and in most cases it is under voluntary control. |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9BE40 | MYH1 | Myosin-1 | Bos taurus (Bovine) | SS |
| Q9BE41 | MYH2 | Myosin-2 | Bos taurus (Bovine) | SS |
| Q27991 | MYH10 | Myosin-10 | Bos taurus (Bovine) | SS |
| Q9BE39 | MYH7 | Myosin-7 | Bos taurus (Bovine) | SS |
| P10587 | MYH11 | Myosin-11 | Gallus gallus (Chicken) | SS |
| P14105 | MYH9 | Myosin-9 | Gallus gallus (Chicken) | SS |
| P13538 | Myosin heavy chain, skeletal muscle, adult | Gallus gallus (Chicken) | SS | |
| P02565 | MYH1B | Myosin-1B | Gallus gallus (Chicken) | SS |
| Q99323 | zip | Myosin heavy chain, non-muscle | Drosophila melanogaster (Fruit fly) | SS |
| P05661 | Mhc | Myosin heavy chain, muscle | Drosophila melanogaster (Fruit fly) | SS |
| A7E2Y1 | MYH7B | Myosin-7B | Homo sapiens (Human) | SS |
| P11055 | MYH3 | Myosin-3 | Homo sapiens (Human) | SS |
| P12882 | MYH1 | Myosin-1 | Homo sapiens (Human) | SS |
| P12883 | MYH7 | Myosin-7 | Homo sapiens (Human) | EV |
| P13533 | MYH6 | Myosin-6 | Homo sapiens (Human) | SS |
| Q9UKX3 | MYH13 | Myosin-13 | Homo sapiens (Human) | SS |
| Q9Y2K3 | MYH15 | Myosin-15 | Homo sapiens (Human) | SS |
| Q9Y623 | MYH4 | Myosin-4 | Homo sapiens (Human) | SS |
| Q9UKX2 | MYH2 | Myosin-2 | Homo sapiens (Human) | SS |
| P35580 | MYH10 | Myosin-10 | Homo sapiens (Human) | SS |
| P35749 | MYH11 | Myosin-11 | Homo sapiens (Human) | SS |
| P35579 | MYH9 | Myosin-9 | Homo sapiens (Human) | SS |
| Q7Z406 | MYH14 | Myosin-14 | Homo sapiens (Human) | SS |
| Q8VDD5 | Myh9 | Myosin-9 | Mus musculus (Mouse) | SS |
| Q5SX39 | Myh4 | Myosin-4 | Mus musculus (Mouse) | SS |
| Q02566 | Myh6 | Myosin-6 | Mus musculus (Mouse) | SS |
| O08638 | Myh11 | Myosin-11 | Mus musculus (Mouse) | SS |
| A2AQP0 | Myh7b | Myosin-7B | Mus musculus (Mouse) | SS |
| Q61879 | Myh10 | Myosin-10 | Mus musculus (Mouse) | SS |
| Q91Z83 | Myh7 | Myosin-7 | Mus musculus (Mouse) | SS |
| Q6URW6 | Myh14 | Myosin-14 | Mus musculus (Mouse) | SS |
| P13541 | Myh3 | Myosin-3 | Mus musculus (Mouse) | SS |
| Q5SX40 | Myh1 | Myosin-1 | Mus musculus (Mouse) | SS |
| P13542 | Myh8 | Myosin-8 | Mus musculus (Mouse) | SS |
| P79293 | MYH7 | Myosin-7 | Sus scrofa (Pig) | SS |
| Q9TV63 | MYH2 | Myosin-2 | Sus scrofa (Pig) | SS |
| P12847 | Myh3 | Myosin-3 | Rattus norvegicus (Rat) | SS |
| P02563 | Myh6 | Myosin-6 | Rattus norvegicus (Rat) | SS |
| P02564 | Myh7 | Myosin-7 | Rattus norvegicus (Rat) | SS |
| Q62812 | Myh9 | Myosin-9 | Rattus norvegicus (Rat) | SS |
| Q29RW1 | Myh4 | Myosin-4 | Rattus norvegicus (Rat) | SS |
| Q9JLT0 | Myh10 | Myosin-10 | Rattus norvegicus (Rat) | SS |
| P02566 | unc-54 | Myosin-4 | Caenorhabditis elegans | SS |
| P02567 | myo-1 | Myosin-1 | Caenorhabditis elegans | SS |
| P12844 | myo-3 | Myosin-3 | Caenorhabditis elegans | SS |
| P12845 | myo-2 | Myosin-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSASSDAEMA | VFGEAAPYLR | KSEKERIEAQ | NKPFDAKTSV | FVAEPKESYV | KSTIQSKEGG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KVTVKTEGGA | TLTVREDQVF | PMNPPKYDKI | EDMAMMTHLH | EPGVLYNLKE | RYAAWMIYTY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SGLFCVTVNP | YKWLPVYKPE | VVAAYRGKKR | QEAPPHIFSI | SDNAYQFMLT | DRENQSILIT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GESGAGKTVN | TKRVIQYFAT | IAVTGEKKKD | ESGKMQGTLE | DQIISANPLL | EAFGNAKTVR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NDNSSRFGKF | IRIHFGTTGK | LASADIETYL | LEKSRVTFQL | KAERSYHIFY | QITSNKKPDL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IEMLLITTNP | YDYAFVSQGE | ITVPSIDDQE | ELMATDSAID | ILGFTPEEKV | SIYKLTGAVM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| HYGNMKFKQK | QREEQAEPDG | TEVADKAAYL | QSLNSADLLK | ALCYPRVKVG | NEYVTKGQTV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QQVYNAVGAL | AKAVYEKMFL | WMVTRINQQL | DTKQPRQYFI | GVLDIAGFEI | FDFNSLEQLC |
| 490 | 500 | 510 | 520 | 530 | 540 |
| INFTNEKLQQ | FFNHHMFVLE | QEEYKKEGIE | WTFIDFGMDL | AACIELIEKP | LGIFSILEEE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| CMFPKATDTS | FKNKLYDQHL | GKSANFQKPK | VVKGKAEAHF | SLIHYAGTVD | YNITGWLDKN |
| 610 | 620 | 630 | 640 | 650 | 660 |
| KDPLNDTVVG | LYQKSAMKTL | ASLFSTYASA | EADSSAKKGA | KKKGSSFQTV | SALFRENLNK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LMTNLRSTHP | HFVRCIIPNE | TKTPGAMEHE | LVLHQLRCNG | VLEGIRICRK | GFPSRILYGD |
| 730 | 740 | 750 | 760 | 770 | 780 |
| FKQRYKVLNA | SAIPEGQFID | SKKASEKLLA | SIDIDHTQYK | FGHTKVFFKA | GLLGLLEEMR |
| 790 | 800 | 810 | 820 | 830 | 840 |
| DEKLAQIITR | TQAVCRGFLM | RVEYQKMLQR | REALFCIQYN | VRAFMNVKHW | PWMKLFFKIK |
| 850 | 860 | 870 | 880 | 890 | 900 |
| PLLKSAETEK | EMATMKEEFQ | KTKDELAKSE | AKRKELEEKM | VTLLKEKNDL | QLQVQSEADS |
| 910 | 920 | 930 | 940 | 950 | 960 |
| LADAEERCEQ | LIKNKIQLEA | KIKEVTERAE | EEEEINAELT | AKKRKLEDEC | SELKKDIDDL |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| ELTLAKVEKE | KHATENKVKN | LTEEMAGLDE | TIAKLSKEKK | ALQETHQQTL | DDLQAEEDKV |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| NILTKAKTKL | EQQVDDLEGS | LEQEKKLRMD | LERAKRKLEG | DLKLAQESTM | DMENDKQQLD |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| EKLEKKEFEI | SNLISKIEDE | QAVEIQLQKK | IKELQARIEE | LGEEIEAERA | SRAKAEKQRS |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| DLSRELEEIS | ERLEEAGGAT | SAQVELNKKR | EAEFQKLRRD | LEEATLQHEA | MVAALRKKHA |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| DSMAELGEQI | DNLQRVKQKL | EKEKSELKME | TDDLSSNAEA | ISKAKGNLEK | MCRSLEDQVS |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| ELKTKEEEQQ | RLINDLTAQR | ARLQTEAGEY | SRQLDEKDAL | VSQLSRSKQA | STQQIEELKH |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| QLEEETKAKN | ALAHALQSSR | HDCDLLREQY | EEEQEGKAEL | QRALSKANSE | VAQWRTKYET |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| DAIQRTEELE | EAKKKLAQRL | QEAEEHVEAV | NAKCASLEKT | KQRLQNEVED | LMLDVERSNA |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| ACAALDKKQR | NFDKVLSEWK | QKYEETQAEL | EASQKESRSL | STELFKVKNV | YEESLDQLET |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| LRRENKNLQQ | EISDLTEQIA | EGGKQIHELE | KIKKQVEQEK | CEIQAALEEA | EASLEHEEGK |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| ILRIQLELNQ | VKSEVDRKIA | EKDEEIDQLK | RNHTRVVETM | QSTLDAEIRS | RNDALRVKKK |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| MEGDLNEMEI | QLNHANRLAA | ESLRNYRNTQ | GILKETQLHL | DDALRGQEDL | KEQLAIVERR |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| ANLLQAEIEE | LWATLEQTER | SRKIAEQELL | DASERVQLLH | TQNTSLINTK | KKLENDVSQL |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| QSEVEEVIQE | SRNAEEKAKK | AITDAAMMAE | ELKKEQDTSA | HLERMKKNLE | QTVKDLQHRL |
| 1810 | 1820 | 1830 | 1840 | 1850 | 1860 |
| DEAEQLALKG | GKKQIQKLEA | RVRELEGEVE | NEQKRNAEAV | KGLRKHERRV | KELTYQTEED |
| 1870 | 1880 | 1890 | 1900 | 1910 | 1920 |
| RKNVLRLQDL | VDKLQAKVKS | YKRQAEEAEE | QSNANLSKFR | KLQHELEEAE | ERADIAESQV |
| 1930 | |||||
| NKLRVKSREV | HTKISAE |