P11801
Gene name |
PSKH1 |
Protein name |
Serine/threonine-protein kinase H1 |
Names |
EC 2.7.11.1 , Protein serine kinase H1 , PSK-H1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5681 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
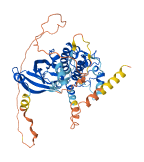
Descriptions
Autoinhibitory domains (AIDs)
Accessory elements
238-262 (Activation loop from InterPro)
Target domain |
98-355 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
Autoinhibited structure

Activated structure

1 structures for P11801
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P11801-F1 | Predicted | AlphaFoldDB |
382 variants for P11801
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1269561499 | 2 | G>D | No | gnomAD | |
| rs764221259 | 5 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs764221259 | 5 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs763298582 | 5 | T>P | No | ExAC | |
| rs774585594 | 6 | S>N | No |
ExAC gnomAD |
|
| rs1250866121 | 7 | K>N | No |
TOPMed gnomAD |
|
| rs2058163591 | 8 | V>A | No | Ensembl | |
| rs889100595 | 8 | V>I | No |
TOPMed gnomAD |
|
| rs760619786 | 9 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs766401988 | 9 | L>P | No |
ExAC gnomAD |
|
| rs764767985 | 11 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs2058163686 | 12 | P>L | No | gnomAD | |
| rs758072788 | 12 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1273419897 | 13 | P>S | No |
TOPMed gnomAD |
|
| rs777211025 | 14 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs976552733 | 15 | D>G | No | TOPMed | |
| rs1598189210 | 15 | D>N | No | TOPMed | |
| rs138557834 | 16 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs533725307 | 19 | D>V | No | Ensembl | |
| rs1174221423 | 24 | V>M | No |
TOPMed gnomAD |
|
| rs1223803260 | 26 | P>L | No | gnomAD | |
| rs2058163843 | 26 | P>T | No |
TOPMed gnomAD |
|
| rs2058163876 | 28 | S>I | No | Ensembl | |
| rs781752881 | 29 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs781752881 | 29 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs770154201 | 30 | T>I | No |
ExAC gnomAD |
|
| rs1567399158 | 30 | T>P | No | Ensembl | |
| rs1236774193 | 31 | K>N | No | gnomAD | |
| rs780302290 | 31 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1481013467 | 32 | S>G | No | gnomAD | |
| rs749464076 | 32 | S>I | No |
ExAC gnomAD |
|
| TCGA novel | 32 | S>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769003520 | 33 | D>N | No |
ExAC gnomAD |
|
| rs762038809 | 34 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs374100286 | 37 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1356246890 | 40 | T>I | No | gnomAD | |
| rs759426626 | 42 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2058164101 | 43 | D>N | No | TOPMed | |
| rs2058164130 | 44 | S>I | No | TOPMed | |
| rs576450048 | 45 | V>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs146186709 | 46 | G>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs146186709 | 46 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs146186709 | 46 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs763803391 | 47 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1283454159 | 50 | A>V | No | gnomAD | |
| rs201663239 | 51 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs750891126 | 52 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs979745952 | 52 | F>S | No | Ensembl | |
| rs1207190109 | 53 | P>L | No | gnomAD | |
| rs2058164341 | 53 | P>S | No | Ensembl | |
|
rs780398848 COSM3888787 |
54 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1482769126 | 55 | A>G | No |
TOPMed gnomAD |
|
| rs2058164407 | 57 | Q>* | No | Ensembl | |
| rs372174162 | 58 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1598189309 | 60 | H>P | No | Ensembl | |
| rs1339969747 | 60 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs545099030 | 61 | P>S | No | Ensembl | |
| rs1313538872 | 63 | P>S | No | TOPMed | |
| rs1056019516 | 64 | G>D | No |
TOPMed gnomAD |
|
| rs139742716 | 64 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1056019516 | 64 | G>V | No |
TOPMed gnomAD |
|
| rs560568634 | 65 | P>S | No | gnomAD | |
| rs560568634 | 65 | P>T | No | gnomAD | |
| rs144509198 | 66 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs781187782 | 67 | T>P | No |
ExAC TOPMed gnomAD |
|
| COSM3361855 | 69 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1417999141 | 70 | H>Q | No | TOPMed | |
| rs2058164706 | 70 | H>R | No | TOPMed | |
| rs1788857944 | 71 | T>A | No |
TOPMed gnomAD |
|
| rs753168298 | 71 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1567399224 | 73 | P>R | No | Ensembl | |
| TCGA novel | 74 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM435551 | 75 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2058164824 | 76 | E>G | No | TOPMed | |
| rs1230609617 | 77 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1236245758 | 77 | P>S | No | TOPMed | |
| rs2058164859 | 78 | P>L | No |
TOPMed gnomAD |
|
| rs751254936 | 79 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs1341567923 COSM972614 |
79 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1341567923 | 79 | R>L | No |
TOPMed gnomAD |
|
| rs1275498288 | 80 | R>K | No |
TOPMed gnomAD |
|
| rs2058164967 | 81 | A>V | No | Ensembl | |
| rs1217090998 | 82 | R>G | No |
TOPMed gnomAD |
|
| rs1317491228 | 84 | A>T | No | TOPMed | |
| rs761392649 | 85 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs936187440 | 86 | Y>C | No | TOPMed | |
| rs113404164 | 89 | K>E | No | Ensembl | |
| rs756639302 | 89 | K>R | No |
ExAC gnomAD |
|
| rs767047171 | 93 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs754228505 COSM4062004 |
93 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2058165140 | 94 | V>I | No | TOPMed | |
| rs764660707 | 98 | Y>* | No |
ExAC gnomAD |
|
| rs754273417 | 99 | D>V | No |
ExAC gnomAD |
|
| rs748355795 | 100 | I>S | No | ExAC | |
| rs2058165258 | 101 | K>* | No |
TOPMed gnomAD |
|
| rs1345573875 | 102 | A>V | No | gnomAD | |
| rs747128704 | 106 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs775455575 | 111 | R>* | No |
ExAC gnomAD |
|
| rs775455575 | 111 | R>G | No |
ExAC gnomAD |
|
| rs1332605943 | 111 | R>Q | No | gnomAD | |
| rs1598189418 | 112 | V>G | No | Ensembl | |
| rs1209473224 | 114 | R>C | No |
TOPMed gnomAD |
|
| rs768402377 | 114 | R>H | No |
ExAC gnomAD |
|
| rs1209473224 | 114 | R>S | No |
TOPMed gnomAD |
|
| rs774143232 | 115 | V>I | No |
ExAC gnomAD |
|
| rs1598189427 | 117 | H>P | No | Ensembl | |
| rs2058165528 | 118 | R>Q | No | gnomAD | |
| rs55913417 | 118 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1445820858 | 119 | A>V | No |
TOPMed gnomAD |
|
| rs1598189440 | 120 | T>P | No | Ensembl | |
| rs760148388 | 121 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs772761696 | 121 | R>W | No |
ExAC TOPMed gnomAD |
|
|
rs754355030 COSM1609537 |
123 | P>L | liver [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs2151312349 | 123 | P>T | No | Ensembl | |
| rs1598189453 | 124 | Y>D | No | Ensembl | |
| rs1458455792 | 124 | Y>F | No |
TOPMed gnomAD |
|
| rs151268578 | 126 | I>M | No |
ESP TOPMed gnomAD |
|
| rs1318348259 | 126 | I>V | No |
TOPMed gnomAD |
|
| rs1453369763 | 127 | K>Q | No | gnomAD | |
| rs1201861827 | 128 | M>I | No | gnomAD | |
| rs1338789283 | 129 | I>T | No | gnomAD | |
| rs2058165786 | 131 | T>I | No | TOPMed | |
| rs540279720 | 133 | Y>* | No |
ExAC gnomAD |
|
| rs530075963 | 133 | Y>C | No |
ExAC gnomAD |
|
| rs777914939 | 134 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs570571625 | 134 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777914939 | 134 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs563460830 | 136 | G>W | No |
ExAC gnomAD |
|
| rs1462044574 | 137 | R>G | No |
TOPMed gnomAD |
|
| rs1441582991 | 137 | R>Q | No | gnomAD | |
|
COSM1222312 rs1462044574 |
137 | R>W | large_intestine [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1598189487 | 139 | V>G | No | Ensembl | |
| TCGA novel | 141 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369110023 | 144 | L>M | No |
ESP TOPMed gnomAD |
|
| rs749163092 | 145 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs768583092 | 145 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2058165997 | 146 | V>L | No | TOPMed | |
| rs531259214 | 148 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs771730022 | 148 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1306879769 | 149 | R>L | No |
TOPMed gnomAD |
|
| rs1306879769 | 149 | R>Q | No |
TOPMed gnomAD |
|
| rs373245459 | 149 | R>W | No |
ESP ExAC gnomAD |
|
| rs1598189515 | 150 | V>G | No | Ensembl | |
|
rs1399930911 COSM4062006 |
151 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1019592266 | 151 | R>H | No |
TOPMed gnomAD |
|
| rs2151312374 | 153 | A>S | No | Ensembl | |
| rs2058166140 | 154 | N>S | No | Ensembl | |
| rs765915108 | 156 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs753096981 | 157 | Q>H | No |
ExAC gnomAD |
|
| rs377334046 | 159 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1598189535 | 161 | V>G | No | Ensembl | |
| rs2058166240 | 163 | E>K | No | gnomAD | |
|
rs751693739 COSM1709301 |
165 | Q>R | skin [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1701677720 | 166 | E>D | No | gnomAD | |
| rs2058166269 | 166 | E>G | No | gnomAD | |
| rs781180501 | 167 | R>Q | No |
ExAC gnomAD |
|
| rs757495547 | 167 | R>W | No |
ExAC gnomAD |
|
| rs1598189547 | 168 | V>G | No | Ensembl | |
| rs1483711142 | 168 | V>M | No | gnomAD | |
| rs201098098 | 170 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs201098098 | 170 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1598189553 | 171 | V>G | No | Ensembl | |
| rs1433588898 | 173 | E>D | No | gnomAD | |
| rs771721386 | 175 | A>G | No |
ExAC gnomAD |
|
| rs747940857 | 175 | A>T | No |
ExAC gnomAD |
|
| rs2058166390 | 177 | G>D | No | gnomAD | |
| TCGA novel | 178 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746605543 | 178 | G>V | No |
ExAC gnomAD |
|
| rs535771946 | 179 | E>A | No |
1000Genomes ExAC gnomAD |
|
| rs776280871 | 183 | R>C | No |
ExAC gnomAD |
|
| rs141391249 | 183 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1598189570 | 186 | A>T | No | Ensembl | |
| COSM972615 | 187 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1388714909 | 188 | G>S | No | gnomAD | |
| rs376814861 | 192 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs775902065 | 192 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1277318997 | 193 | R>C | No |
TOPMed gnomAD |
|
| rs764318654 | 193 | R>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 194 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2058166508 | 195 | A>T | No | Ensembl | |
| rs1598189583 | 195 | A>V | No | Ensembl | |
| rs1273243169 | 196 | T>M | No | gnomAD | |
| rs767784614 | 197 | R>Q | No |
ExAC gnomAD |
|
| rs147652625 | 197 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1432318624 | 198 | V>L | No | gnomAD | |
| rs1432318624 | 198 | V>M | No | gnomAD | |
| rs752525986 | 200 | Q>R | No |
ExAC gnomAD |
|
| rs777367014 | 206 | V>D | No |
ExAC gnomAD |
|
| rs777367014 | 206 | V>G | No |
ExAC gnomAD |
|
| rs2058166645 | 206 | V>L | No | TOPMed | |
| rs770694440 | 207 | R>Q | No |
ExAC gnomAD |
|
| rs746869313 | 207 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs781041385 | 212 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs745344209 | 217 | R>* | No |
ExAC gnomAD |
|
| rs2058166754 | 217 | R>P | No | gnomAD | |
| COSM972616 | 217 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1055067510 | 218 | D>N | No |
TOPMed gnomAD |
|
| rs1289006170 | 225 | L>F | No | TOPMed | |
| rs763313027 | 226 | Y>H | No |
ExAC gnomAD |
|
| rs2058166839 | 227 | Y>D | No | Ensembl | |
| rs2151312443 | 228 | H>Y | No | Ensembl | |
| TCGA novel | 229 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1463531263 | 229 | P>L | No |
TOPMed gnomAD |
|
| rs2058166850 | 229 | P>T | No | TOPMed | |
| TCGA novel | 230 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1446385916 | 231 | T>A | No | gnomAD | |
| rs1316650317 | 231 | T>I | No | Ensembl | |
| rs774682036 | 234 | K>E | No |
ExAC gnomAD |
|
| rs762209921 | 235 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs773383066 | 236 | I>T | No |
ExAC gnomAD |
|
| rs767690823 | 236 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs368564776 | 243 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs143148420 | 246 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs751378850 | 246 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs751378850 | 246 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs757163361 | 247 | K>E | No |
ExAC gnomAD |
|
| COSM4756863 | 247 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs931071569 | 247 | K>R | No |
TOPMed gnomAD |
|
| rs2058167048 | 248 | K>M | No | Ensembl | |
| rs2058167036 | 248 | K>Q | No | Ensembl | |
| rs781168612 | 249 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs148210457 | 249 | G>S | No | ESP | |
| rs781168612 | 249 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs745472725 | 252 | C>R | No |
ExAC gnomAD |
|
| rs1214996799 | 252 | C>Y | No | gnomAD | |
| rs1484294432 | 256 | T>S | No |
TOPMed gnomAD |
|
| rs1598189668 | 257 | T>P | No | Ensembl | |
| rs2058167138 | 257 | T>S | No | TOPMed | |
|
COSM4062008 rs1180232058 |
260 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1473074727 | 262 | E>A | No | gnomAD | |
| rs1348475152 | 263 | Y>S | No |
TOPMed gnomAD |
|
| rs192629945 | 264 | I>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs192629945 | 264 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs919782915 | 265 | A>P | No | Ensembl | |
| rs1008475522 | 268 | V>I | No | TOPMed | |
| rs774874464 | 270 | V>A | No |
ExAC gnomAD |
|
| rs748685487 | 271 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs772373019 | 271 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs772373019 | 271 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1453355214 | 273 | P>L | No |
TOPMed gnomAD |
|
| rs1410057570 | 273 | P>T | No | gnomAD | |
| rs760740011 | 276 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs766692223 | 277 | S>L | No |
ExAC gnomAD |
|
| rs751505949 | 280 | M>I | No |
ExAC gnomAD |
|
| rs759710609 | 280 | M>L | No | ExAC | |
| rs759710609 | 280 | M>V | No | ExAC | |
| rs2058167393 | 281 | W>* | No | Ensembl | |
| rs2058167404 | 282 | A>T | No | TOPMed | |
| rs2058167411 | 282 | A>V | No | Ensembl | |
| COSM703811 | 283 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4926272 | 284 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201009210 | 285 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1483385552 | 287 | A>V | No | gnomAD | |
| rs1017219009 | 292 | S>C | No |
TOPMed gnomAD |
|
| rs1017219009 | 292 | S>G | No |
TOPMed gnomAD |
|
| rs778417363 | 294 | T>S | No |
ExAC gnomAD |
|
| rs1374332593 | 295 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1429770607 | 296 | P>L | No | gnomAD | |
|
VAR_040614 COSM3818304 rs35552721 |
301 | N>S | breast [Cosmic] | No |
cosmic curated UniProt ExAC TOPMed dbSNP gnomAD |
| rs1275281711 | 302 | R>C | No |
TOPMed gnomAD |
|
|
COSM1222313 rs747466620 |
302 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1275281711 | 302 | R>S | No |
TOPMed gnomAD |
|
| rs368590968 | 303 | T>I | No | Ensembl | |
| rs1279451077 | 304 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1217412354 | 304 | R>W | No | gnomAD | |
| rs776952921 | 307 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs776952921 | 307 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs771036863 | 307 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1387936268 | 308 | Q>H | No |
TOPMed gnomAD |
|
| rs1598189745 | 309 | I>V | No | Ensembl | |
| rs1022063287 | 310 | L>I | No |
TOPMed gnomAD |
|
| rs769949236 | 311 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs2058167746 | 313 | K>N | No | TOPMed | |
| rs2058167740 | 313 | K>R | No |
TOPMed gnomAD |
|
| rs775728687 | 314 | Y>D | No |
ExAC TOPMed gnomAD |
|
| rs775728687 | 314 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs767617738 | 315 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs750159369 | 315 | S>N | No |
ExAC gnomAD |
|
| rs750159369 | 315 | S>T | No |
ExAC gnomAD |
|
| rs1598189768 | 317 | S>C | No | Ensembl | |
| rs760554211 | 318 | G>R | No |
ExAC gnomAD |
|
| rs760554211 | 318 | G>W | No |
ExAC gnomAD |
|
| TCGA novel | 319 | E>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1252053815 | 323 | S>G | No |
TOPMed gnomAD |
|
| rs749548353 | 330 | D>N | No |
ExAC gnomAD |
|
| rs2058220147 | 332 | I>N | No | TOPMed | |
| rs183508494 | 334 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs377532074 | 334 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs2058220255 | 338 | V>M | No | Ensembl | |
| rs2058220265 | 342 | A>V | No | Ensembl | |
| rs770838620 | 343 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs770838620 | 343 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1308905185 | 343 | R>H | No | gnomAD | |
| rs1308905185 | 343 | R>L | No | gnomAD | |
| COSM4062009 | 343 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1214099248 | 345 | T>S | No |
TOPMed gnomAD |
|
| COSM972617 | 346 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774453539 | 347 | L>P | No | Ensembl | |
| rs1598195256 | 352 | H>P | No | Ensembl | |
| rs1352594835 | 353 | P>L | No |
TOPMed gnomAD |
|
| rs202036975 | 354 | W>* | No | 1000Genomes | |
| rs1598195270 | 355 | V>G | No | Ensembl | |
| rs1306749423 | 355 | V>M | No | gnomAD | |
| rs761912701 | 356 | V>L | No | Ensembl | |
| rs1232398074 | 357 | S>R | No |
TOPMed gnomAD |
|
| rs1236153522 | 358 | M>L | No |
TOPMed gnomAD |
|
| rs1567402817 | 358 | M>R | No | Ensembl | |
| rs1567402817 | 358 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1598195282 | 359 | A>P | No | Ensembl | |
| rs1273563995 | 360 | A>D | No |
TOPMed gnomAD |
|
| rs1273563995 | 360 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| COSM972618 | 361 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2058220460 | 363 | S>F | No | gnomAD | |
| rs764840386 | 364 | M>V | No |
ExAC gnomAD |
|
| TCGA novel | 367 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4062010 rs199995143 |
369 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs199995143 | 369 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM972619 rs763604843 |
369 | R>H | large_intestine endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs199995143 | 369 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1296576921 | 370 | S>F | No | TOPMed | |
| rs1249639695 | 371 | I>V | No |
TOPMed gnomAD |
|
| rs1567402832 | 373 | Q>L | No | Ensembl | |
| rs141214862 | 375 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs145190865 | 378 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs202152584 | 378 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 379 | A>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 379 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756503993 | 380 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs137887537 | 381 | S>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201909376 | 382 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs575605513 | 382 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1008919267 | 383 | C>Y | No | TOPMed | |
| rs2058220681 | 386 | T>A | No | Ensembl | |
| rs1324257769 | 389 | A>T | No | gnomAD | |
| rs770877782 | 389 | A>V | No | ExAC | |
| rs766265181 | 390 | Q>H | No |
TOPMed gnomAD |
|
| rs745836876 | 392 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs561480493 | 393 | R>C | No |
1000Genomes TOPMed gnomAD |
|
|
rs201432888 COSM972620 |
393 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs775281500 | 394 | S>P | No |
ExAC gnomAD |
|
| rs1721410480 | 395 | S>G | No | gnomAD | |
| rs867603325 | 395 | S>I | No |
TOPMed gnomAD |
|
| rs755016530 | 396 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs778967004 | 396 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs778967004 | 396 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs773901928 | 398 | T>I | No |
ExAC gnomAD |
|
| rs1467986549 | 398 | T>S | No | gnomAD | |
|
COSM972621 rs1000942376 |
399 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs968616577 | 399 | R>H | No |
TOPMed gnomAD |
|
| rs2058220936 | 401 | N>D | No | TOPMed | |
| rs376293494 | 401 | N>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs376293494 | 401 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1333866869 | 402 | K>R | No |
TOPMed gnomAD |
|
| rs2058220992 | 403 | S>P | No | Ensembl | |
| rs756698298 | 404 | R>C | No |
ExAC gnomAD |
|
| rs766782692 | 404 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs754320519 | 405 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs755212982 | 405 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs2058221074 | 406 | V>A | No | Ensembl | |
| rs779051799 | 406 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs752646115 | 407 | R>Q | No |
TOPMed gnomAD |
|
|
COSM3691113 rs748229359 |
407 | R>W | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1294185088 | 408 | E>K | No | gnomAD | |
| rs1234681997 | 409 | R>Q | No |
TOPMed gnomAD |
|
| rs146152043 | 409 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1567402915 | 410 | E>K | No | TOPMed | |
| rs1567402915 | 410 | E>Q | No | TOPMed | |
| rs745748731 | 412 | R>Q | No |
ExAC gnomAD |
|
| rs777852511 | 412 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1186493255 | 413 | E>* | No | gnomAD | |
| rs1186493255 | 413 | E>K | No | gnomAD | |
| rs1209336836 | 414 | L>F | No | gnomAD | |
| rs2058221259 | 415 | N>H | No | TOPMed | |
| rs1484831785 | 415 | N>S | No | gnomAD | |
| rs1186030654 | 417 | R>C | No |
TOPMed gnomAD |
|
| rs1256886493 | 417 | R>H | No |
TOPMed gnomAD |
|
| rs1425788462 | 419 | Q>R | No | gnomAD | |
| rs199646456 | 421 | Q>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs986396947 | 422 | Y>* | No |
TOPMed gnomAD |
|
| rs915631938 | 423 | N>H | No | Ensembl | |
| rs1381152492 | 423 | N>S | No | gnomAD | |
| rs1343980301 | 424 | G>S | No | gnomAD |
No associated diseases with P11801
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| nuclear speck | A discrete extra-nucleolar subnuclear domain, 20-50 in number, in which splicing factors are seen to be localized by immunofluorescence microscopy. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| determination of left/right symmetry | The establishment of an organism's body plan or part of an organism with respect to the left and right halves. The pattern can either be symmetric, such that the halves are mirror images, or asymmetric where the pattern deviates from this symmetry. |
| heart development | The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
106 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4IFM7 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Bos taurus (Bovine) | SS |
| Q0VD22 | STK33 | Serine/threonine-protein kinase 33 | Bos taurus (Bovine) | PR |
| Q0V7M1 | PSKH1 | Serine/threonine-protein kinase H1 | Bos taurus (Bovine) | SS |
| O15075 | DCLK1 | Serine/threonine-protein kinase DCLK1 | Homo sapiens (Human) | EV |
| Q9UIK4 | DAPK2 | Death-associated protein kinase 2 | Homo sapiens (Human) | EV |
| O94768 | STK17B | Serine/threonine-protein kinase 17B | Homo sapiens (Human) | PR |
| Q16566 | CAMK4 | Calcium/calmodulin-dependent protein kinase type IV | Homo sapiens (Human) | SS |
| Q8N568 | DCLK2 | Serine/threonine-protein kinase DCLK2 | Homo sapiens (Human) | PR |
| Q9UEE5 | STK17A | Serine/threonine-protein kinase 17A | Homo sapiens (Human) | SS |
| O75962 | TRIO | Triple functional domain protein | Homo sapiens (Human) | EV |
| Q8WWF8 | CAPSL | Calcyphosin-like protein | Homo sapiens (Human) | PR |
| P53355 | DAPK1 | Death-associated protein kinase 1 | Homo sapiens (Human) | EV |
| Q9BYT3 | STK33 | Serine/threonine-protein kinase 33 | Homo sapiens (Human) | PR |
| O43293 | DAPK3 | Death-associated protein kinase 3 | Homo sapiens (Human) | PR |
| Q14012 | CAMK1 | Calcium/calmodulin-dependent protein kinase type 1 | Homo sapiens (Human) | EV |
| Q9H1R3 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Homo sapiens (Human) | EV |
| Q16816 | PHKG1 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform | Homo sapiens (Human) | PR |
| P15735 | PHKG2 | Phosphorylase b kinase gamma catalytic chain, liver/testis isoform | Homo sapiens (Human) | PR |
| Q13554 | CAMK2B | Calcium/calmodulin-dependent protein kinase type II subunit beta | Homo sapiens (Human) | EV |
| Q9UQM7 | CAMK2A | Calcium/calmodulin-dependent protein kinase type II subunit alpha | Homo sapiens (Human) | EV |
| Q13555 | CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | Homo sapiens (Human) | EV |
| Q13557 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Homo sapiens (Human) | EV |
| Q32MK0 | MYLK3 | Myosin light chain kinase 3 | Homo sapiens (Human) | SS |
| Q86YV6 | MYLK4 | Myosin light chain kinase family member 4 | Homo sapiens (Human) | SS |
| Q6P2M8 | PNCK | Calcium/calmodulin-dependent protein kinase type 1B | Homo sapiens (Human) | SS |
| Q96NX5 | CAMK1G | Calcium/calmodulin-dependent protein kinase type 1G | Homo sapiens (Human) | SS |
| Q8IU85 | CAMK1D | Calcium/calmodulin-dependent protein kinase type 1D | Homo sapiens (Human) | SS |
| Q8NCB2 | CAMKV | CaM kinase-like vesicle-associated protein | Homo sapiens (Human) | SS |
| Q9QYK9 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Mus musculus (Mouse) | PR |
| Q91VB2 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Mus musculus (Mouse) | SS |
| Q8BG48 | Stk17b | Serine/threonine-protein kinase 17B | Mus musculus (Mouse) | PR |
| Q8VCR8 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Mus musculus (Mouse) | SS |
| Q6PGN3 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Mus musculus (Mouse) | PR |
| Q91YS8 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Mus musculus (Mouse) | SS |
| Q3UIZ8 | Mylk3 | Myosin light chain kinase 3 | Mus musculus (Mouse) | SS |
| Q8VDF3 | Dapk2 | Death-associated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q6P8Y1 | Capsl | Calcyphosin-like protein | Mus musculus (Mouse) | PR |
| Q8BW96 | Camk1d | Calcium/calmodulin-dependent protein kinase type 1D | Mus musculus (Mouse) | SS |
| Q3UHL1 | Camkv | CaM kinase-like vesicle-associated protein | Mus musculus (Mouse) | SS |
| P08414 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Mus musculus (Mouse) | SS |
| Q9JLM8 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Mus musculus (Mouse) | SS |
| Q80YE7 | Dapk1 | Death-associated protein kinase 1 | Mus musculus (Mouse) | SS |
| Q62407 | Speg | Striated muscle-specific serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| Q0KL02 | Trio | Triple functional domain protein | Mus musculus (Mouse) | SS |
| Q924X7 | Stk33 | Serine/threonine-protein kinase 33 | Mus musculus (Mouse) | PR |
| O54784 | Dapk3 | Death-associated protein kinase 3 | Mus musculus (Mouse) | PR |
| Q91YA2 | Pskh1 | Serine/threonine-protein kinase H1 | Mus musculus (Mouse) | SS |
| Q7TNJ7 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Rattus norvegicus (Rat) | SS |
| P20689 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Rattus norvegicus (Rat) | SS |
| F1M0Z1 | Trio | Triple functional domain protein | Rattus norvegicus (Rat) | SS |
| Q91XS8 | Stk17b | Serine/threonine-protein kinase 17B | Rattus norvegicus (Rat) | PR |
| O08875 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Rattus norvegicus (Rat) | PR |
| E9PT87 | Mylk3 | Myosin light chain kinase 3 | Rattus norvegicus (Rat) | SS |
| O70150 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Rattus norvegicus (Rat) | PR |
| Q63092 | Camkv | CaM kinase-like vesicle-associated protein | Rattus norvegicus (Rat) | SS |
| Q63638 | Speg | Striated muscle-specific serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| O88764 | Dapk3 | Death-associated protein kinase 3 | Rattus norvegicus (Rat) | PR |
| P13234 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Rattus norvegicus (Rat) | SS |
| Q63450 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Rattus norvegicus (Rat) | EV |
| P97924 | Kalrn | Kalirin | Rattus norvegicus (Rat) | SS |
| Q5MPA9 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Rattus norvegicus (Rat) | PR |
| Q6AVM3 | CCAMK | Calcium and calcium/calmodulin-dependent serine/threonine-protein kinase | Oryza sativa subsp japonica (Rice) | PR |
| P53682 | CPK23 | Calcium-dependent protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| P53684 | CPK7 | Calcium-dependent protein kinase 7 | Oryza sativa subsp japonica (Rice) | PR |
| Q5VQQ5 | CPK2 | Calcium-dependent protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6I5I8 | CPK16 | Calcium-dependent protein kinase 16 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6K968 | CPK6 | Calcium-dependent protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| Q75GE8 | CPK8 | Calcium-dependent protein kinase 8 | Oryza sativa subsp. japonica (Rice) | SS |
| Q84SL0 | CPK20 | Calcium-dependent protein kinase 20 | Oryza sativa subsp. japonica (Rice) | SS |
| Q8LPZ7 | CPK3 | Calcium-dependent protein kinase 3 | Oryza sativa subsp. japonica (Rice) | SS |
| Q9TXJ0 | cmk-1 | Calcium/calmodulin-dependent protein kinase type 1 | Caenorhabditis elegans | PR |
| O44997 | dapk-1 | Death-associated protein kinase dapk-1 | Caenorhabditis elegans | SS |
| Q95QC4 | zyg-8 | Serine/threonine-protein kinase zyg-8 | Caenorhabditis elegans | PR |
| P28583 | Calcium-dependent protein kinase SK5 | Glycine max (Soybean) (Glycine hispida) | PR | |
| Q9M9V8 | CPK10 | Calcium-dependent protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FX86 | CRK8 | CDPK-related kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SSF8 | CPK30 | Calcium-dependent protein kinase 30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIQ7 | CPK24 | Calcium-dependent protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93759 | GK-1 | Calcium-dependent protein kinase 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8S8S2 | LPEAT2 | Lysophospholipid acyltransferase LPEAT2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LJL9 | CRK2 | CDPK-related kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SG12 | CRK6 | CDPK-related kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCS2 | CRK5 | CDPK-related kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W4I7 | CPK13 | Calcium-dependent protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NLQ6 | CPK32 | Calcium-dependent protein kinase 32 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38873 | CPK7 | Calcium-dependent protein kinase 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42438 | CPK8 | Calcium-dependent protein kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q06850 | CPK1 | Calcium-dependent protein kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38869 | CPK4 | Calcium-dependent protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38870 | CPK2 | Calcium-dependent protein kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38872 | CPK6 | Calcium-dependent protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39016 | CPK11 | Calcium-dependent protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E9C0 | CPK34 | Calcium-dependent protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42396 | CPK12 | Calcium-dependent protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42479 | CPK3 | Calcium-dependent protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7XJR9 | CPK16 | Calcium-dependent protein kinase 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FMP5 | CPK17 | Calcium-dependent protein kinase 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJ61 | CPK25 | Calcium-dependent protein kinase 25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZSA4 | CPK27 | Calcium-dependent protein kinase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZV15 | CPK20 | Calcium-dependent protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1PE17 | CPK18 | Calcium-dependent protein kinase 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FKW4 | CPK28 | Calcium-dependent protein kinase 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1LUA6 | trio | Triple functional domain protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A8C984 | mylk3 | Myosin light chain kinase 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q7SY49 | camkv | CaM kinase-like vesicle-associated protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGCGTSKVLP | EPPKDVQLDL | VKKVEPFSGT | KSDVYKHFIT | EVDSVGPVKA | GFPAASQYAH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PCPGPPTAGH | TEPPSEPPRR | ARVAKYRAKF | DPRVTAKYDI | KALIGRGSFS | RVVRVEHRAT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RQPYAIKMIE | TKYREGREVC | ESELRVLRRV | RHANIIQLVE | VFETQERVYM | VMELATGGEL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FDRIIAKGSF | TERDATRVLQ | MVLDGVRYLH | ALGITHRDLK | PENLLYYHPG | TDSKIIITDF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GLASARKKGD | DCLMKTTCGT | PEYIAPEVLV | RKPYTNSVDM | WALGVIAYIL | LSGTMPFEDD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NRTRLYRQIL | RGKYSYSGEP | WPSVSNLAKD | FIDRLLTVDP | GARMTALQAL | RHPWVVSMAA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SSSMKNLHRS | ISQNLLKRAS | SRCQSTKSAQ | STRSSRSTRS | NKSRRVRERE | LRELNLRYQQ |
| QYNG |