P11799
Gene name |
Mylk |
Protein name |
Myosin light chain kinase, smooth muscle |
Names |
MLCK , EC 2.7.11.18 , Telokin [Cleaved into: Myosin light chain kinase, smooth muscle, deglutamylated form] |
Species |
Gallus gallus (Chicken) |
KEGG Pathway |
|
EC number |
2.7.11.18: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1453-1708 (Protein kinase domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
1593-1615 (Activation loop from InterPro)
Target domain |
1453-1708 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
1593-1615 (Activation loop from InterPro)
Target domain |
1450-1708 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
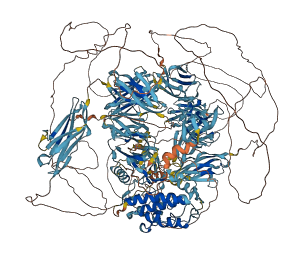
Autoinhibited structure

Activated structure

16 structures for P11799
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1CDL | X-ray | 200 A | E/F/G/H | 1730-1749 | PDB |
| 1QS7 | X-ray | 180 A | B/D | 1731-1749 | PDB |
| 1QTX | X-ray | 165 A | B | 1731-1749 | PDB |
| 1VRK | X-ray | 190 A | B | 1731-1749 | PDB |
| 2O5G | X-ray | 108 A | B | 1730-1748 | PDB |
| 3EK7 | X-ray | 185 A | A | 1730-1763 | PDB |
| 3EK8 | X-ray | 280 A | A | 1730-1763 | PDB |
| 3EKH | X-ray | 200 A | A | 1730-1763 | PDB |
| 3EKJ | X-ray | 280 A | A | 1730-1763 | PDB |
| 3EVR | X-ray | 200 A | A | 1730-1763 | PDB |
| 3EVU | X-ray | 175 A | A | 1731-1749 | PDB |
| 3EVV | X-ray | 260 A | A | 1731-1749 | PDB |
| 3O77 | X-ray | 235 A | A | 1730-1749 | PDB |
| 3O78 | X-ray | 260 A | A/B | 1731-1749 | PDB |
| 4OY4 | X-ray | 203 A | PDB | ||
| AF-P11799-F1 | Predicted | AlphaFoldDB |
No variants for P11799
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P11799 | |||||
No associated diseases with P11799
41 regional properties for P11799
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 1453 - 1708 | IPR000719 |
| domain | Immunoglobulin subtype 2 | 40 - 108 | IPR003598-1 |
| domain | Immunoglobulin subtype 2 | 168 - 235 | IPR003598-2 |
| domain | Immunoglobulin subtype 2 | 441 - 508 | IPR003598-3 |
| domain | Immunoglobulin subtype 2 | 540 - 604 | IPR003598-4 |
| domain | Immunoglobulin subtype 2 | 649 - 716 | IPR003598-5 |
| domain | Immunoglobulin subtype 2 | 747 - 814 | IPR003598-6 |
| domain | Immunoglobulin subtype 2 | 1096 - 1163 | IPR003598-7 |
| domain | Immunoglobulin subtype 2 | 1237 - 1304 | IPR003598-8 |
| domain | Immunoglobulin subtype 2 | 1806 - 1874 | IPR003598-9 |
| domain | Immunoglobulin subtype | 34 - 119 | IPR003599-1 |
| domain | Immunoglobulin subtype | 162 - 246 | IPR003599-2 |
| domain | Immunoglobulin subtype | 435 - 519 | IPR003599-3 |
| domain | Immunoglobulin subtype | 534 - 615 | IPR003599-4 |
| domain | Immunoglobulin subtype | 643 - 727 | IPR003599-5 |
| domain | Immunoglobulin subtype | 741 - 825 | IPR003599-6 |
| domain | Immunoglobulin subtype | 1090 - 1174 | IPR003599-7 |
| domain | Immunoglobulin subtype | 1231 - 1315 | IPR003599-8 |
| domain | Immunoglobulin subtype | 1800 - 1885 | IPR003599-9 |
| domain | Fibronectin type III | 1318 - 1414 | IPR003961 |
| domain | Immunoglobulin-like domain | 28 - 117 | IPR007110-1 |
| domain | Immunoglobulin-like domain | 156 - 244 | IPR007110-2 |
| domain | Immunoglobulin-like domain | 429 - 517 | IPR007110-3 |
| domain | Immunoglobulin-like domain | 521 - 613 | IPR007110-4 |
| domain | Immunoglobulin-like domain | 637 - 725 | IPR007110-5 |
| domain | Immunoglobulin-like domain | 735 - 830 | IPR007110-6 |
| domain | Immunoglobulin-like domain | 1084 - 1160 | IPR007110-7 |
| domain | Immunoglobulin-like domain | 1225 - 1313 | IPR007110-8 |
| domain | Immunoglobulin-like domain | 1794 - 1885 | IPR007110-9 |
| active_site | Serine/threonine-protein kinase, active site | 1570 - 1582 | IPR008271 |
| domain | Immunoglobulin I-set | 28 - 118 | IPR013098-1 |
| domain | Immunoglobulin I-set | 156 - 245 | IPR013098-2 |
| domain | Immunoglobulin I-set | 429 - 518 | IPR013098-3 |
| domain | Immunoglobulin I-set | 529 - 614 | IPR013098-4 |
| domain | Immunoglobulin I-set | 639 - 726 | IPR013098-5 |
| domain | Immunoglobulin I-set | 736 - 822 | IPR013098-6 |
| domain | Immunoglobulin I-set | 1084 - 1173 | IPR013098-7 |
| domain | Immunoglobulin I-set | 1225 - 1314 | IPR013098-8 |
| domain | Immunoglobulin I-set | 1794 - 1884 | IPR013098-9 |
| domain | Myosin Light Chain Kinase 1, Kinase domain | 1450 - 1708 | IPR015725 |
| binding_site | Protein kinase, ATP binding site | 1459 - 1482 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.18 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cleavage furrow | The cleavage furrow is a plasma membrane invagination at the cell division site. The cleavage furrow begins as a shallow groove and eventually deepens to divide the cytoplasm. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| stress fiber | A contractile actin filament bundle that consists of short actin filaments with alternating polarity, cross-linked by alpha-actinin and possibly other actin bundling proteins, and with myosin present in a periodic distribution along the fiber. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| metal ion binding | Binding to a metal ion. |
| myosin light chain kinase activity | Catalysis of the reaction |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| muscle structure development | The progression of a muscle structure over time, from its formation to its mature state. Muscle structures are contractile cells, tissues or organs that are found in multicellular organisms. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| tonic smooth muscle contraction | A process in which force is generated within tonic smooth muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis. In the tonic smooth muscle, the muscle contraction occurs without an ordered sarcomeric structure. Tonic smooth muscle contraction occurs as a sustained continuous contraction. |
9 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8WZ42 | TTN | Titin | Homo sapiens (Human) | EV |
| Q15746 | MYLK | Myosin light chain kinase, smooth muscle | Homo sapiens (Human) | SS |
| Q62407 | Speg | Striated muscle-specific serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| A2ASS6 | Ttn | Titin | Mus musculus (Mouse) | SS |
| Q6PDN3 | Mylk | Myosin light chain kinase, smooth muscle | Mus musculus (Mouse) | EV |
| Q63638 | Speg | Striated muscle-specific serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| P97924 | Kalrn | Kalirin | Rattus norvegicus (Rat) | SS |
| G4SLH0 | ttn-1 | Titin homolog | Caenorhabditis elegans | EV |
| Q23551 | unc-22 | Twitchin | Caenorhabditis elegans | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGDVKLVTST | RVSKTSLTLS | PSVPAEAPAF | TLPPRNIRVQ | LGATARFEGK | VRGYPEPQIT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| WYRNGHPLPE | GDHYVVDHSI | RGIFSLVIKG | VQEGDSGKYT | CEAANDGGVR | QVTVELTVEG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NSLKKYSLPS | SAKTPGGRLS | VPPVEHRPSI | WGESPPKFAT | KPNRVVVREG | QTGRFSCKIT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GRPQPQVTWT | KGDIHLQQNE | RFNMFEKTGI | QYLEIQNVQL | ADAGIYTCTV | VNSAGKASVS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AELTVQGPDK | TDTHAQPLCM | PPKPTTLATK | AIENSDFKQA | TSNGIAKELK | STSTELMVET |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KDRLSAKKET | FYTSREAKDG | KQGQNQEANA | VPLQESRGTK | GPQVLQKTSS | TITLQAVKAQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PEPKAEPQTT | FIRQAEDRKR | TVQPLMTTTT | QENPSLTGQV | SPRSRETENR | AGVRKSVKEE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KREPLGIPPQ | FESRPQSLEA | SEGQEIKFKS | KVSGKPKPDV | EWFKEGVPIK | TGEGIQIYEE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DGTHCLWLKK | ACLGDSGSYS | CAAFNPRGQT | STSWLLTVKR | PKVEEVAPCF | SSVLKGCTVS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EGQDFVLQCY | VGGVPVPEIT | WLLNEQPIQY | AHSTFEAGVA | KLTVQDALPE | DDGIYTCLAE |
| 610 | 620 | 630 | 640 | 650 | 660 |
| NNAGRASCSA | QVTVKEKKSS | KKAEGTQAAK | LNKTFAPIFL | KGLTDLKVMD | GSQVIMTVEV |
| 670 | 680 | 690 | 700 | 710 | 720 |
| SANPCPEIIW | LHNGKEIQET | EDFHFEKKGN | EYSLYIQEVF | PEDTGKYTCE | AWNELGETQT |
| 730 | 740 | 750 | 760 | 770 | 780 |
| QATLTVQEPQ | DGIQPWFISK | PRSVTAAAGQ | NVLISCAIAG | DPFPTVHWFK | DGQEITPGTG |
| 790 | 800 | 810 | 820 | 830 | 840 |
| CEILQNEDIF | TLILRNVQSR | HAGQYEIQLR | NQVGECSCQV | SLMLRESSAS | RAEMLRDGRE |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SASSGERRDG | GNYGALTFGR | TSGFKKSSSE | TRAAEEEQED | VRGVLKRRVE | TREHTEESLR |
| 910 | 920 | 930 | 940 | 950 | 960 |
| QQEAEQLDFR | DILGKKVSTK | SFSEEDLKEI | PAEQMDFRAN | LQRQVKPKTL | SEEERKVHAP |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| QQVDFRSVLA | KKGTPKTPLP | EKVPPPKPAV | TDFRSVLGAK | KKPPAENGSA | STPAPNARAG |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| SEAQNATPNS | EAPAPKPVVK | KEEKNDRKCE | HGCAVVDGGI | IGKKAENKPA | ASKPTPPPSK |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| GTAPSFTEKL | QDAKVADGEK | LVLQCRISSD | PPASVSWTLD | SKAIKSSKSI | VISQEGTLCS |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| LTIEKVMPED | GGEYKCIAEN | AAGKAECACK | VLVEDTSSTK | AAKPAEKKTK | KPKTTLPPVL |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| STESSEATVK | KKPAPKTPPK | AATPPQITQF | PEDRKVRAGE | SVELFAKVVG | TAPITCTWMK |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| FRKQIQENEY | IKIENAENSS | KLTISSTKQE | HCGCYTLVVE | NKLGSRQAQV | NLTVVDKPDP |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| PAGTPCASDI | RSSSLTLSWY | GSSYDGGSAV | QSYTVEIWNS | VDNKWTDLTT | CRSTSFNVQD |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| LQADREYKFR | VRAANVYGIS | EPSQESEVVK | VGEKQEEELK | EEEAELSDDE | GKETEVNYRT |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| VTINTEQKVS | DVYNIEERLG | SGKFGQVFRL | VEKKTGKVWA | GKFFKAYSAK | EKENIRDEIS |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| IMNCLHHPKL | VQCVDAFEEK | ANIVMVLEMV | SGGELFERII | DEDFELTERE | CIKYMRQISE |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| GVEYIHKQGI | VHLDLKPENI | MCVNKTGTSI | KLIDFGLARR | LESAGSLKVL | FGTPEFVAPE |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| VINYEPIGYE | TDMWSIGVIC | YILVSGLSPF | MGDNDNETLA | NVTSATWDFD | DEAFDEISDD |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| AKDFISNLLK | KDMKSRLNCT | QCLQHPWLQK | DTKNMEAKKL | SKDRMKKYMA | RRKWQKTGHA |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| VRAIGRLSSM | AMISGMSGRK | ASGSSPTSPI | NADKVENEDA | FLEEVAEEKP | HVKPYFTKTI |
| 1810 | 1820 | 1830 | 1840 | 1850 | 1860 |
| LDMEVVEGSA | ARFDCKIEGY | PDPEVMWYKD | DQPVKESRHF | QIDYDEEGNC | SLTISEVCGD |
| 1870 | 1880 | 1890 | 1900 | ||
| DDAKYTCKAV | NSLGEATCTA | ELLVETMGKE | GEGEGEGEED | EEEEEE |