P11488
Gene name |
GNAT1 (GNATR) |
Protein name |
Guanine nucleotide-binding protein G |
Names |
t subunit alpha-1 , Transducin alpha-1 chain |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2779 |
EC number |
|
Protein Class |
|
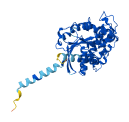
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
180-344 (Ras-like domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Goricanec D et al. (2016) "Conformational dynamics of a G-protein α subunit is tightly regulated by nucleotide binding", Proceedings of the National Academy of Sciences of the United States of America, 113, E3629-38
- Coleman DE et al. (1999) "Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex", The Journal of biological chemistry, 274, 16669-72
- Lutz S et al. (2007) "Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs", Science (New York, N.Y.), 318, 1923-7
Autoinhibited structure

Activated structure

2 structures for P11488
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3RBQ | X-ray | 200 A | G/H/I/J/K/L | 2-11 | PDB |
| AF-P11488-F1 | Predicted | AlphaFoldDB |
430 variants for P11488
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1575415957 RCV001003035 |
3 | A>missing | Retinitis pigmentosa [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001412336 RCV000299430 CA2412421 rs201006405 |
13 | R>K | Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000259457 CA2412433 rs149936603 RCV001437241 |
28 | R>Q | Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001370367 rs772024349 RCV000787836 |
36 | G>D | Retinitis pigmentosa Retinitis pigmentosa (rp) [ClinVar, Ensembl] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000017277 CA126053 VAR_009279 rs104893740 |
38 | G>D | Congenital stationary night blindness autosomal dominant 3 CSNBAD3 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt ExAC dbSNP gnomAD |
|
RCV002050635 rs765432703 RCV003355558 |
79 | A>G | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs201849628 RCV001498016 CA2412489 RCV000361876 RCV002520163 |
81 | V>I | Inborn genetic diseases Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001197099 rs1699444080 |
87 | L>F | Congenital stationary night blindness 1G [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001388651 RCV002476734 rs143481438 |
91 | Y>* | Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001075209 rs778059585 |
95 | A>missing | Retinal dystrophy [ClinVar] | Yes |
ClinVar dbSNP |
|
rs778497200 RCV001593238 RCV001060334 |
120 | S>* | Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV003166747 RCV001306731 rs199611280 |
125 | R>Q | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002573468 RCV001971465 rs540227694 |
125 | R>W | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV000171141 VAR_073798 rs786205854 CA250324 |
129 | D>G | Congenital stationary night blindness 1G CSNB1G [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs771466852 RCV001347341 RCV002547075 |
148 | A>S | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001003036 rs1293620319 |
154 | D>Y | Retinitis pigmentosa [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
rs1559746613 RCV000785165 RCV000785164 |
163 | Y>S | Congenital stationary night blindness 1G Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs149647295 RCV001724867 |
170 | V>L | Retinitis pigmentosa Retinitis pigmentosa (rp) [ClinVar, Ensembl] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
CA199760 RCV000171140 rs786205853 VAR_073799 |
200 | Q>E | Congenital stationary night blindness autosomal dominant 3 CSNBAD3 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs777846284 RCV003246877 RCV001321257 |
204 | R>G | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
RCV001233414 rs769077570 RCV001150686 |
258 | T>M | Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs751585772 RCV001351190 RCV002547518 |
270 | F>L | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs886058685 CA10616962 RCV000383999 |
277 | A>V | Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
RCV000578484 RCV001237857 RCV001150687 CA2412765 rs374913800 |
302 | Q>* | Congenital stationary night blindness 1G Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000382489 CA2412769 RCV001850834 rs367790137 |
309 | R>P | Congenital stationary night blindness autosomal dominant 3 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs772111717 RCV001073737 |
316 | Y>C | Retinal dystrophy [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
rs1699480267 RCV001199362 |
339 | I>V | Congenital stationary night blindness 1G [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001348235 rs753375778 |
1 | M>I | No |
ClinVar dbSNP |
|
|
RCV001968871 rs763736641 |
1 | M>V | No |
ClinVar dbSNP |
|
| rs1420138504 | 2 | G>V | No |
TOPMed gnomAD |
|
|
rs1266812415 RCV002001578 |
3 | A>T | No |
ClinVar TOPMed dbSNP |
|
|
rs1699413898 RCV001867666 |
4 | G>R | No |
ClinVar Ensembl dbSNP |
|
| rs1699413898 | 4 | G>W | No | Ensembl | |
|
rs780667765 RCV001317593 |
6 | S>G | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs1459356965 | 6 | S>R | No |
TOPMed gnomAD |
|
|
rs1293986442 COSM1424120 |
7 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 8 | E>D | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1384503071 | 8 | E>Q | No | gnomAD | |
| rs1575415985 | 9 | E>A | No | Ensembl | |
|
RCV000082616 rs398124522 CA224144 |
9 | E>missing | No |
ClinGen ClinVar dbSNP |
|
| COSM1424121 | 10 | K>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1377171417 | 11 | H>Y | No | gnomAD | |
| rs748389570 | 14 | E>D | No |
ExAC TOPMed gnomAD |
|
| COSM5583542 | 14 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770031393 | 16 | E>G | No |
ExAC gnomAD |
|
| rs1699414664 | 17 | K>N | No | TOPMed | |
|
rs753761683 RCV001883391 |
18 | K>missing | No |
ClinVar dbSNP |
|
| rs1699414848 | 20 | K>E | No | Ensembl | |
| rs1177827474 | 23 | A>D | No | gnomAD | |
| rs1053549443 | 23 | A>S | No |
TOPMed gnomAD |
|
| rs1053549443 | 23 | A>T | No |
TOPMed gnomAD |
|
| rs1378270615 | 25 | K>N | No |
TOPMed gnomAD |
|
| rs1575416010 | 25 | K>R | No | Ensembl | |
| rs1425289444 | 26 | D>E | No | gnomAD | |
| rs1161256318 | 27 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1165407105 | 27 | A>V | No | gnomAD | |
|
rs774214573 RCV000658963 COSM3595519 |
28 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ClinVar ExAC NCI-TCGA dbSNP gnomAD |
| rs149936603 | 28 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1400630067 | 29 | T>A | No | gnomAD | |
| rs145040990 | 30 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs145040990 RCV001878192 |
30 | V>M | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs2109137515 RCV002025294 |
34 | L>H | No |
ClinVar Ensembl dbSNP |
|
| rs1487464362 | 36 | G>S | No |
TOPMed gnomAD |
|
| TCGA novel | 38 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1347967126 | 38 | G>S | No | gnomAD | |
|
rs34797487 RCV001307134 |
39 | E>D | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1575416566 | 40 | S>P | No | Ensembl | |
| rs1699439597 | 41 | G>A | No | Ensembl | |
| rs768429545 | 41 | G>W | No | ExAC | |
| rs1699439647 | 42 | K>E | No |
TOPMed gnomAD |
|
| rs776367777 | 42 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1575416575 | 44 | T>P | No | Ensembl | |
|
rs764910362 RCV002020824 |
46 | V>I | No |
ClinVar ExAC dbSNP gnomAD |
|
| COSM116906 | 47 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs749921670 RCV001910851 |
48 | Q>E | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs1559746052 | 49 | M>I | No | Ensembl | |
| rs1466539542 | 49 | M>V | No | TOPMed | |
|
rs2109138350 RCV001930469 |
50 | K>missing | No |
ClinVar dbSNP |
|
| rs1699441764 | 53 | H>R | No | TOPMed | |
| rs778486199 | 54 | Q>* | No | Ensembl | |
| rs776566245 | 55 | D>E | No |
ExAC TOPMed gnomAD |
|
|
rs1041904269 RCV001305259 |
55 | D>Y | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs190126440 | 57 | Y>* | No |
1000Genomes TOPMed gnomAD |
|
| rs1242103172 | 60 | E>K | No |
TOPMed gnomAD |
|
| rs1242103172 | 60 | E>Q | No |
TOPMed gnomAD |
|
| rs1699442559 | 63 | L>V | No | Ensembl | |
| rs1352647422 | 65 | F>L | No | gnomAD | |
| rs762489106 | 70 | Y>* | No |
ExAC TOPMed gnomAD |
|
|
rs767935708 RCV001373234 |
71 | G>C | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs767935708 | 71 | G>S | No |
ExAC gnomAD |
|
| rs776116147 | 72 | N>H | No |
ExAC gnomAD |
|
| rs1236654107 | 73 | T>K | No | TOPMed | |
|
rs1236654107 RCV001209348 |
73 | T>M | No |
ClinVar TOPMed dbSNP |
|
| rs1699443183 | 75 | Q>* | No | Ensembl | |
| COSM269252 | 75 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV001961109 rs764511104 |
77 | I>F | No |
ClinVar ExAC dbSNP gnomAD |
|
|
RCV001314060 rs754189877 |
77 | I>M | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs757483692 | 78 | L>Q | No |
ExAC gnomAD |
|
| rs765432703 | 79 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs1699443540 | 80 | I>M | No | TOPMed | |
| rs1699443661 | 81 | V>A | No |
TOPMed gnomAD |
|
| rs201849628 | 81 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1406092062 | 82 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1699443746 | 82 | R>H | No | TOPMed | |
| rs1406092062 | 82 | R>S | No |
TOPMed gnomAD |
|
|
RCV002040640 COSM145454 rs1325267297 |
83 | A>T | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ClinVar NCI-TCGA TOPMed dbSNP gnomAD |
| rs1559746197 | 83 | A>V | No | Ensembl | |
| TCGA novel | 84 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 85 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1575416692 | 85 | T>P | No | Ensembl | |
|
rs2109138520 RCV001382720 |
86 | T>missing | No |
ClinVar dbSNP |
|
| rs1338199628 | 86 | T>A | No |
TOPMed gnomAD |
|
| rs2109138528 | 86 | T>K | No | Ensembl | |
| rs1338199628 | 86 | T>P | No |
TOPMed gnomAD |
|
| rs1338199628 | 86 | T>S | No |
TOPMed gnomAD |
|
|
RCV002009853 rs767518257 |
88 | N>S | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1375880116 | 89 | I>L | No | gnomAD | |
| rs1055779105 | 90 | Q>* | No | Ensembl | |
| COSM1424122 | 90 | Q>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV001244594 rs1403539761 |
91 | Y>C | No |
ClinVar Ensembl dbSNP |
|
| rs747889085 | 92 | G>R | No |
ExAC gnomAD |
|
|
rs772876090 RCV001219562 |
93 | D>G | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs2109138554 RCV001903560 |
93 | D>I | No |
ClinVar Ensembl dbSNP |
|
| rs769366813 | 93 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs772876090 | 93 | D>V | No |
ExAC TOPMed gnomAD |
|
| COSM1046424 | 94 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1424123 | 96 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs748872282 | 96 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs748872282 | 96 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1418453394 | 96 | R>S | No |
TOPMed gnomAD |
|
| rs1263559950 | 98 | D>N | No |
TOPMed gnomAD |
|
| rs1263559950 | 98 | D>Y | No |
TOPMed gnomAD |
|
| rs766450007 | 99 | D>A | No |
ExAC gnomAD |
|
| rs759644439 | 100 | A>T | No |
ExAC gnomAD |
|
|
COSM1046426 rs767686983 |
101 | R>Q | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1222830660 | 103 | L>V | No |
TOPMed gnomAD |
|
| rs1014696655 | 104 | M>R | No | TOPMed | |
|
RCV001316043 rs1699447851 |
104 | M>V | No |
ClinVar Ensembl dbSNP |
|
| rs965873488 | 105 | H>Y | No |
TOPMed gnomAD |
|
| rs1699448058 | 106 | M>I | No | gnomAD | |
| rs752725817 | 106 | M>L | No |
ExAC gnomAD |
|
| rs918866339 | 106 | M>T | No | Ensembl | |
| rs752906907 | 107 | A>V | No |
TOPMed gnomAD |
|
| rs2109138728 | 109 | T>S | No | Ensembl | |
|
RCV001226202 rs756003441 |
110 | I>N | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs777683622 | 112 | E>A | No |
ExAC gnomAD |
|
| rs753519744 | 113 | G>R | No |
ExAC gnomAD |
|
| rs1279193204 | 114 | T>M | No |
TOPMed gnomAD |
|
| rs2109138755 | 119 | M>I | No | Ensembl | |
|
rs778497200 COSM4118712 |
120 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs778497200 | 120 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs2109138765 | 121 | D>H | No | 1000Genomes | |
| rs1453636003 | 122 | I>S | No | gnomAD | |
| rs771618545 | 122 | I>V | No |
ExAC gnomAD |
|
| rs200450293 | 123 | I>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1699448634 | 123 | I>N | No | TOPMed | |
| rs200450293 | 123 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2109138776 | 124 | Q>* | No | Ensembl | |
| rs748754383 | 124 | Q>R | No |
ExAC gnomAD |
|
| TCGA novel | 125 | R>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs199611280 | 125 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
|
rs763156863 RCV001045234 |
126 | L>Q | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs759804674 | 127 | W>* | No |
ExAC gnomAD |
|
| rs373544213 | 127 | W>G | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001723414 rs373544213 |
127 | W>R | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs928804929 RCV001948880 |
130 | S>A | No |
ClinVar Ensembl dbSNP |
|
| rs752708194 | 131 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
RCV001033959 rs141497735 |
132 | I>S | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1240433262 | 133 | Q>* | No | gnomAD | |
| TCGA novel | 133 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1055894672 | 133 | Q>P | No | Ensembl | |
|
RCV001301799 rs1699452379 |
134 | A>G | No |
ClinVar Ensembl dbSNP |
|
| rs1699452452 | 135 | C>Y | No | TOPMed | |
| rs753698158 | 136 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs138533270 | 137 | E>K | No |
ESP ExAC |
|
|
RCV001989206 rs1268403135 |
138 | R>G | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs2109138833 | 138 | R>H | No | Ensembl | |
| rs758039880 | 139 | A>P | No |
ExAC gnomAD |
|
| rs758039880 | 139 | A>S | No |
ExAC gnomAD |
|
| COSM2851060 | 139 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1440003996 | 140 | S>* | No |
TOPMed gnomAD |
|
| rs1361979499 | 141 | E>* | No |
TOPMed gnomAD |
|
| rs1453708491 | 141 | E>G | No | gnomAD | |
| rs1361979499 | 141 | E>K | No |
TOPMed gnomAD |
|
| TCGA novel | 141 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778098995 | 145 | N>I | No |
ExAC gnomAD |
|
| rs1699453394 | 146 | D>E | No | TOPMed | |
| rs1699453366 | 146 | D>N | No | TOPMed | |
| rs771466852 | 148 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 148 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746153500 | 149 | G>C | No |
ExAC gnomAD |
|
| rs1306986926 | 149 | G>D | No |
TOPMed gnomAD |
|
| rs746153500 | 149 | G>S | No |
ExAC gnomAD |
|
| rs1306986926 | 149 | G>V | No |
TOPMed gnomAD |
|
| rs375063932 | 151 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs752200483 | 151 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1699457258 | 151 | Y>N | No | TOPMed | |
| rs1293620319 | 154 | D>H | No |
TOPMed gnomAD |
|
|
rs1293620319 RCV001995962 |
154 | D>N | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1279604764 | 155 | L>P | No | gnomAD | |
| rs1211321547 | 155 | L>V | No |
TOPMed gnomAD |
|
| rs779522093 | 157 | R>C | No |
ExAC gnomAD |
|
| rs779522093 | 157 | R>G | No |
ExAC gnomAD |
|
| rs1334509024 | 157 | R>L | No | Ensembl | |
| rs779522093 | 157 | R>S | No |
ExAC gnomAD |
|
| rs1242217198 | 159 | V>I | No |
TOPMed gnomAD |
|
| TCGA novel | 160 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1474755659 | 160 | T>N | No |
TOPMed gnomAD |
|
| rs1474755659 | 160 | T>S | No |
TOPMed gnomAD |
|
| rs1399876988 | 161 | P>A | No |
TOPMed gnomAD |
|
| rs1699458313 | 161 | P>L | No | TOPMed | |
|
rs1399876988 RCV002027438 |
161 | P>T | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs995607401 | 162 | G>A | No | Ensembl | |
| rs747259508 | 163 | Y>* | No |
ExAC gnomAD |
|
| rs1455904263 | 164 | V>A | No |
TOPMed gnomAD |
|
| rs1455904263 | 164 | V>E | No |
TOPMed gnomAD |
|
| rs1405575787 | 164 | V>M | No |
TOPMed gnomAD |
|
| rs781142235 | 165 | P>R | No |
ExAC gnomAD |
|
| rs768908387 | 165 | P>S | No |
ExAC gnomAD |
|
| rs1028826170 | 166 | T>I | No |
TOPMed gnomAD |
|
| rs1439988252 | 166 | T>S | No | TOPMed | |
| rs1028826170 | 166 | T>S | No |
TOPMed gnomAD |
|
|
rs769750483 RCV001883932 |
167 | E>* | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs749642690 | 167 | E>D | No | TOPMed | |
| rs769750483 | 167 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs773081995 | 168 | Q>R | No |
ExAC gnomAD |
|
| rs1470513134 | 169 | D>E | No | gnomAD | |
| rs762850401 | 169 | D>H | No |
ExAC gnomAD |
|
|
COSM1424125 rs149647295 RCV002049351 |
170 | V>M | large_intestine Retinitis pigmentosa (rp) [Cosmic, Ensembl] | No |
cosmic curated ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs2109139123 RCV001359694 |
171 | L>Q | No |
ClinVar Ensembl dbSNP |
|
|
COSM1424126 rs1699459195 |
172 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
|
rs1699459195 RCV002276119 |
172 | R>P | No |
ClinVar TOPMed dbSNP |
|
| rs774185121 | 172 | R>S | No |
ExAC gnomAD |
|
| rs914564235 | 173 | S>L | No | TOPMed | |
| rs928754225 | 174 | R>* | No | gnomAD | |
| rs1699459406 | 174 | R>L | No |
TOPMed gnomAD |
|
| COSM4826728 | 174 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1699459521 | 175 | V>A | No | TOPMed | |
| rs760213416 | 175 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1314421649 | 177 | T>I | No | Ensembl | |
| rs1213030549 | 178 | T>I | No | gnomAD | |
| rs1243899846 | 178 | T>S | No | TOPMed | |
| rs1468622659 | 180 | I>N | No |
TOPMed gnomAD |
|
|
RCV001314657 rs751005737 |
181 | I>M | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs758939252 | 182 | E>K | No |
ExAC gnomAD |
|
| rs1452893155 | 183 | T>A | No |
TOPMed gnomAD |
|
|
rs375795574 COSM3781656 |
183 | T>M | Variant assessed as Somatic; MODERATE impact. pancreas [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
RCV001058966 rs1452893155 |
183 | T>P | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1409342994 | 184 | Q>* | No | gnomAD | |
| rs1559746732 | 184 | Q>H | No | Ensembl | |
| rs751891744 | 184 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs1419856584 | 185 | F>I | No | Ensembl | |
| rs1699460164 | 186 | S>Y | No | Ensembl | |
| COSM731215 | 187 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 189 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1272845943 | 189 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| COSM1309296 | 190 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372188249 | 191 | N>K | No |
1000Genomes TOPMed gnomAD |
|
|
RCV002036932 rs1042087188 |
192 | F>L | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs755156826 | 193 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs755156826 | 193 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1699464808 | 194 | M>I | No | Ensembl | |
| rs928625009 | 194 | M>L | No |
TOPMed gnomAD |
|
| rs928625009 | 194 | M>V | No |
TOPMed gnomAD |
|
| rs960398499 | 195 | F>V | No |
TOPMed gnomAD |
|
| rs751898545 | 197 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs755317184 | 199 | G>R | No |
ExAC gnomAD |
|
| rs755317184 | 199 | G>W | No |
ExAC gnomAD |
|
| rs1221785183 | 201 | R>G | No | TOPMed | |
| rs752987189 | 201 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs752987189 | 201 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1699465186 | 202 | S>L | No | TOPMed | |
|
rs571471319 RCV002000220 |
203 | E>* | No |
ClinVar 1000Genomes TOPMed dbSNP gnomAD |
|
| rs777846284 | 204 | R>C | No |
ExAC gnomAD |
|
| rs749325459 | 204 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs749325459 | 204 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs777846284 | 204 | R>S | No |
ExAC gnomAD |
|
| COSM1309297 | 205 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1699465451 | 206 | K>E | No | Ensembl | |
| rs2109139429 | 207 | W>* | No | Ensembl | |
| rs1559746925 | 207 | W>C | No | Ensembl | |
| TCGA novel | 207 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1448085077 | 208 | I>N | No |
TOPMed gnomAD |
|
| rs1699465595 | 209 | H>Y | No | Ensembl | |
| rs2109139440 | 210 | C>F | No | Ensembl | |
| TCGA novel | 211 | F>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745771623 | 212 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs745771623 | 212 | E>Q | No |
ExAC gnomAD |
|
| rs538795841 | 213 | G>D | No |
1000Genomes ExAC gnomAD |
|
| rs1327628053 | 213 | G>S | No | gnomAD | |
|
RCV002003157 rs538795841 |
213 | G>V | No |
ClinVar 1000Genomes ExAC dbSNP gnomAD |
|
| rs1269159422 | 215 | T>A | No |
TOPMed gnomAD |
|
| rs746716339 | 215 | T>I | No |
ExAC gnomAD |
|
| rs1699465851 | 216 | C>Y | No | TOPMed | |
| COSM480246 | 217 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs768364063 RCV001364595 |
218 | I>T | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1486272162 | 219 | F>L | No | gnomAD | |
| rs1699466014 | 219 | F>Y | No | TOPMed | |
| rs1699466127 | 221 | A>T | No | TOPMed | |
| rs766815295 | 222 | A>E | No |
ExAC gnomAD |
|
| rs558297346 | 222 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1372709296 COSM3824210 |
225 | A>T | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
|
RCV001359881 rs2109139492 |
225 | A>V | No |
ClinVar Ensembl dbSNP |
|
| rs534226069 | 226 | Y>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1699466674 | 228 | M>I | No | Ensembl | |
| rs753005762 | 228 | M>V | No |
ExAC gnomAD |
|
| rs1382184490 | 231 | V>A | No | TOPMed | |
| rs764263965 | 231 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1317279783 | 232 | E>A | No | gnomAD | |
| rs1179340527 | 233 | D>H | No |
TOPMed gnomAD |
|
| rs1179340527 | 233 | D>N | No |
TOPMed gnomAD |
|
| rs1226709526 | 234 | D>N | No | gnomAD | |
|
RCV001767606 rs1306113271 |
235 | E>K | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1348995092 | 236 | V>M | No | gnomAD | |
| rs751418123 | 238 | R>H | No |
ExAC gnomAD |
|
| rs751418123 | 238 | R>L | No |
ExAC gnomAD |
|
|
rs1348727426 RCV002003884 |
239 | M>T | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs373605188 | 240 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1699472592 | 241 | E>D | No |
TOPMed gnomAD |
|
| rs1699472630 | 242 | S>R | No | TOPMed | |
| rs780817002 | 243 | L>M | No |
ExAC gnomAD |
|
| rs940268662 | 244 | H>P | No |
TOPMed gnomAD |
|
| rs1699472741 | 245 | L>P | No | TOPMed | |
|
COSM731214 rs1252882654 |
249 | I>V | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs747897846 | 250 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs200018227 | 251 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1476813148 | 252 | H>R | No |
TOPMed gnomAD |
|
| rs777343556 | 253 | R>C | No |
ExAC gnomAD |
|
| rs777343556 | 253 | R>G | No |
ExAC gnomAD |
|
|
rs1475939846 RCV001983314 |
253 | R>H | No |
ClinVar dbSNP gnomAD |
|
| rs748761041 | 254 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs772592889 | 255 | F>L | No |
ExAC gnomAD |
|
| rs772592889 | 255 | F>V | No |
ExAC gnomAD |
|
| rs79816946 | 257 | T>M | No |
ExAC gnomAD |
|
| rs1405282226 | 257 | T>P | No | gnomAD | |
| rs2109139828 | 258 | T>S | No | Ensembl | |
| rs370211781 | 260 | I>V | No | ESP | |
| rs1416751412 | 261 | V>A | No |
TOPMed gnomAD |
|
| rs1416751412 | 261 | V>E | No |
TOPMed gnomAD |
|
| rs1416751412 | 261 | V>G | No |
TOPMed gnomAD |
|
| TCGA novel | 261 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV001315070 rs762106912 |
262 | L>F | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs765309886 | 264 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs763074721 | 268 | D>A | No |
ExAC gnomAD |
|
| rs766541249 | 269 | V>F | No | ExAC | |
| rs766541249 | 269 | V>L | No | ExAC | |
| rs754959065 | 271 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1699474190 | 272 | E>G | No | TOPMed | |
|
RCV001864492 rs1276796249 |
273 | K>missing | No |
ClinVar dbSNP |
|
| rs1486599826 | 273 | K>N | No |
TOPMed gnomAD |
|
|
rs947123026 RCV001903038 |
275 | K>N | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs752370885 | 275 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1481950985 | 276 | K>E | No |
TOPMed gnomAD |
|
| rs1187921368 | 276 | K>M | No | gnomAD | |
| rs886058685 | 277 | A>G | No |
TOPMed gnomAD |
|
| rs532366141 | 278 | H>Y | No |
1000Genomes ExAC gnomAD |
|
| rs187456952 | 279 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs778392995 | 281 | I>V | No |
ExAC gnomAD |
|
|
rs2109139906 RCV002015433 |
285 | D>G | No |
ClinVar Ensembl dbSNP |
|
| rs1699474879 | 287 | D>N | No | Ensembl | |
| rs773704753 | 289 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs954449962 | 290 | N>H | No |
TOPMed gnomAD |
|
| rs749576739 | 290 | N>K | No |
ExAC gnomAD |
|
| rs771213734 | 292 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs759489972 | 293 | E>* | No |
ExAC TOPMed gnomAD |
|
| COSM281413 | 293 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759489972 | 293 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs759489972 | 293 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1013548480 | 294 | D>V | No | Ensembl | |
| rs371673064 | 295 | A>S | No |
ESP ExAC gnomAD |
|
| rs371673064 | 295 | A>T | No |
ESP ExAC gnomAD |
|
|
RCV001300189 rs1241953207 |
296 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinVar NCI-TCGA TOPMed dbSNP gnomAD |
| rs1219121764 | 297 | N>D | No |
TOPMed gnomAD |
|
| rs1699477139 | 297 | N>K | No | TOPMed | |
| rs763936466 | 297 | N>S | No |
ExAC gnomAD |
|
| rs753572984 | 298 | Y>N | No |
ExAC gnomAD |
|
|
RCV001238593 rs772467993 |
299 | I>missing | No |
ClinVar dbSNP |
|
| rs756905728 | 300 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs764827963 | 301 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs757893975 | 302 | Q>R | No |
ExAC gnomAD |
|
| rs779581721 | 304 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1358427033 | 304 | L>H | No | TOPMed | |
| rs779581721 | 304 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs1399146386 | 305 | E>* | No | TOPMed | |
| rs753206978 | 305 | E>G | No |
ExAC gnomAD |
|
| rs924423072 | 308 | M>L | No | Ensembl | |
|
rs1699478168 RCV001302534 |
308 | M>T | No |
ClinVar Ensembl dbSNP |
|
| rs924423072 | 308 | M>V | No | Ensembl | |
| rs367790137 | 309 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs778292500 | 310 | R>C | No |
ExAC gnomAD |
|
| rs1207261147 | 311 | D>E | No | gnomAD | |
|
RCV001048725 rs999064639 |
311 | D>N | No |
ClinVar dbSNP gnomAD |
|
| rs999064639 | 311 | D>Y | No | gnomAD | |
| rs1233953441 | 312 | V>L | No |
TOPMed gnomAD |
|
|
COSM77783 rs1416616268 |
314 | E>D | ovary [Cosmic] | No |
cosmic curated TOPMed |
|
COSM3595520 rs779330892 |
314 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
|
rs746076205 RCV001242161 |
315 | I>V | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs376830581 | 316 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs776440552 | 319 | M>I | No |
ExAC gnomAD |
|
| rs752720192 | 319 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1369841395 | 320 | T>K | No |
TOPMed gnomAD |
|
| rs761545070 | 321 | C>* | No |
ExAC gnomAD |
|
| rs764994417 | 322 | A>V | No |
ExAC gnomAD |
|
| rs1699479544 | 323 | T>A | No |
TOPMed gnomAD |
|
| rs895692880 | 324 | D>N | No |
TOPMed gnomAD |
|
| rs750079496 | 325 | T>M | No |
ExAC gnomAD |
|
| rs750079496 | 325 | T>R | No |
ExAC gnomAD |
|
| rs765861017 | 326 | Q>H | No |
ExAC gnomAD |
|
| rs1440961711 | 326 | Q>K | No | gnomAD | |
| rs1699479769 | 326 | Q>P | No | Ensembl | |
| rs567554388 | 328 | V>L | No |
1000Genomes ExAC gnomAD |
|
| rs202170742 | 329 | K>I | No |
ExAC TOPMed gnomAD |
|
|
RCV001063050 rs202170742 |
329 | K>T | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs754379390 | 332 | F>L | No |
ExAC gnomAD |
|
|
RCV001246958 rs1699480118 |
335 | V>A | No |
ClinVar TOPMed dbSNP |
|
| rs1699480118 | 335 | V>D | No | TOPMed | |
| rs140221218 | 335 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs140221218 | 335 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1366927641 | 336 | T>I | No | gnomAD | |
|
RCV001359238 rs2109140172 |
339 | I>missing | No |
ClinVar dbSNP |
|
| rs1168259545 | 340 | I>S | No | gnomAD | |
| rs1389264153 | 341 | K>R | No |
TOPMed gnomAD |
|
|
TCGA novel rs1699480385 |
342 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| TCGA novel | 342 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780003957 | 342 | E>K | No |
ExAC gnomAD |
|
|
rs747068938 RCV001887667 |
343 | N>S | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs551001437 | 344 | L>V | No | Ensembl | |
| rs1375537782 | 346 | D>G | No | gnomAD | |
| rs1331868461 | 349 | L>H | No | gnomAD | |
| rs1445156543 | 351 | F>L | No |
TOPMed gnomAD |
2 associated diseases with P11488
[MIM: 610444]: Night blindness, congenital stationary, autosomal dominant 3 (CSNBAD3)
A non-progressive retinal disorder characterized by impaired night vision, often associated with nystagmus and myopia. . Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 616389]: Night blindness, congenital stationary, 1G (CSNB1G)
An autosomal recessive form of congenital stationary night blindness, a non-progressive retinal disorder characterized by impaired night vision or in dim light, with good vision only on bright days. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A non-progressive retinal disorder characterized by impaired night vision, often associated with nystagmus and myopia. . Note=The disease is caused by variants affecting the gene represented in this entry.
- An autosomal recessive form of congenital stationary night blindness, a non-progressive retinal disorder characterized by impaired night vision or in dim light, with good vision only on bright days. . Note=The disease is caused by variants affecting the gene represented in this entry.
1 regional properties for P11488
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | MATH/TRAF domain | 7 - 134 | IPR002083 |
Functions
12 GO annotations of cellular component
| Name | Definition |
|---|---|
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| heterotrimeric G-protein complex | Any of a family of heterotrimeric GTP-binding and hydrolyzing proteins; they belong to a superfamily of GTPases that includes monomeric proteins such as EF-Tu and RAS. Heterotrimeric G-proteins consist of three subunits; the alpha subunit contains the guanine nucleotide binding site and possesses GTPase activity; the beta and gamma subunits are tightly associated and function as a beta-gamma heterodimer; extrinsic plasma membrane proteins (cytoplasmic face) that function as a complex to transduce signals from G protein-coupled receptors to an effector protein. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| photoreceptor connecting cilium | The portion of the photoreceptor cell cilium linking the photoreceptor inner and outer segments. It's considered to be equivalent to the ciliary transition zone. |
| photoreceptor disc membrane | Stack of disc membranes located inside a photoreceptor outer segment, and containing densely packed molecules of photoreceptor proteins that traverse the lipid bilayer. Disc membranes arise as evaginations of the ciliary membrane during the development of the outer segment and may or may not remain contiguous with the ciliary membrane. |
| photoreceptor inner segment | The inner segment of a vertebrate photoreceptor containing mitochondria, ribosomes and membranes where opsin molecules are assembled and passed to be part of the outer segment discs. |
| photoreceptor outer segment | The outer segment of a vertebrate photoreceptor that contains a stack of membrane discs embedded with photoreceptor proteins. |
| photoreceptor outer segment membrane | The membrane surrounding the outer segment of a vertebrate photoreceptor. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| acyl binding | Binding to an acyl group, any group formally derived by removal of the hydroxyl group from the acid function of a carboxylic acid. |
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
| metal ion binding | Binding to a metal ion. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
21 GO annotations of biological process
| Name | Definition |
|---|---|
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation or inhibition of adenylyl cyclase activity and a subsequent change in the intracellular concentration of cyclic AMP (cAMP). |
| background adaptation | Any process in which an organism changes its pigmentation (lightening in response to a brighter environment or darkening in response to a dimmer environment) in response to a change in light intensity. |
| cell population proliferation | The multiplication or reproduction of cells, resulting in the expansion of a cell population. |
| cellular response to electrical stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an electrical stimulus. |
| detection of chemical stimulus involved in sensory perception of bitter taste | The series of events required for a bitter taste stimulus to be received and converted to a molecular signal. |
| detection of light stimulus involved in visual perception | The series of events involved in visual perception in which a light stimulus is received and converted into a molecular signal. |
| dopamine secretion | The regulated release of dopamine by a cell. Dopamine is a catecholamine and a precursor of adrenaline and noradrenaline. It acts as a neurotransmitter in the central nervous system but it is also produced peripherally and acts as a hormone. |
| eye photoreceptor cell development | Development of a photoreceptor, a sensory cell in the eye that reacts to the presence of light. They usually contain a pigment that undergoes a chemical change when light is absorbed, thus stimulating a nerve. |
| negative regulation of cyclic-nucleotide phosphodiesterase activity | Any process that stops or reduces the rate of cyclic nucleotide phosphodiesterase activity, the catalysis of the reaction |
| neural tissue regeneration | The regrowth of neural tissue following its loss or destruction. |
| phototransduction, visible light | The sequence of reactions within a cell required to convert absorbed photons from visible light into a molecular signal. A visible light stimulus is electromagnetic radiation that can be perceived visually by an organism; for organisms lacking a visual system, this can be defined as light with a wavelength within the range 380 to 780 nm. |
| regulation of rhodopsin mediated signaling pathway | Any process that modulates the frequency, rate or extent of rhodopsin-mediated signaling. |
| response to light stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a light stimulus, electromagnetic radiation of wavelengths classified as infrared, visible or ultraviolet light. |
| response to organic cyclic compound | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic cyclic compound stimulus. |
| retinal cone cell differentiation | The process in which a relatively unspecialized cell acquires the specialized features of a retinal cone cell. |
| retinal rod cell differentiation | The process in which a relatively unspecialized cell acquires the specialized features of a retinal rod cell. |
| rhodopsin mediated signaling pathway | A G protein-coupled receptor signaling pathway initiated by the excitation of rhodopsin by a photon, and ending with the regulation of a downstream cellular process. |
| sensory perception of umami taste | The series of events required to receive an umami taste stimulus, convert it to a molecular signal, and recognize and characterize the signal. Umami taste is the savory taste of meats and other foods that are rich in glutamates. This is a neurological process. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| visual behavior | The behavior of an organism in response to a visual stimulus. |
| visual perception | The series of events required for an organism to receive a visual stimulus, convert it to a molecular signal, and recognize and characterize the signal. Visual stimuli are detected in the form of photons and are processed to form an image. |
61 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P63097 | GNAI1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P0C7Q4 | GNAT3 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04696 | GNAT2 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P08239 | GNAO1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04695 | GNAT1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P50147 | GNAI2 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P50146 | GNAI1 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P20353 | Galphai | G protein alpha i subunit | Drosophila melanogaster (Fruit fly) | SS |
| P16378 | Galphao | G protein alpha o subunit | Drosophila melanogaster (Fruit fly) | SS |
| P25157 | cta | Guanine nucleotide-binding protein subunit alpha homolog | Drosophila melanogaster (Fruit fly) | SS |
| A8MTJ3 | GNAT3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P08754 | GNAI3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19086 | GNAZ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19087 | GNAT2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P29992 | GNA11 | Guanine nucleotide-binding protein subunit alpha-11 | Homo sapiens (Human) | SS |
| P30679 | GNA15 | Guanine nucleotide-binding protein subunit alpha-15 | Homo sapiens (Human) | SS |
| P50148 | GNAQ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| O95837 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Homo sapiens (Human) | SS |
| Q14344 | GNA13 | Guanine nucleotide-binding protein subunit alpha-13 | Homo sapiens (Human) | SS |
| Q03113 | GNA12 | Guanine nucleotide-binding protein subunit alpha-12 | Homo sapiens (Human) | SS |
| P38405 | GNAL | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P04899 | GNAI2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P63096 | GNAI1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | EV |
| P09471 | GNAO1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q9DC51 | Gnai3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P27600 | Gna12 | Guanine nucleotide-binding protein subunit alpha-12 | Mus musculus (Mouse) | SS |
| P27601 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Mus musculus (Mouse) | SS |
| Q3V3I2 | Gnat3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| O70443 | Gnaz | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P50149 | Gnat2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P08752 | Gnai2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| B2RSH2 | Gnai1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P18872 | Gnao1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P20612 | Gnat1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P93564 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum tuberosum (Potato) | SS |
| Q63210 | Gna12 | Guanine nucleotide-binding protein subunit alpha-12 | Rattus norvegicus (Rat) | SS |
| P19627 | Gnaz | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P29348 | Gnat3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P04897 | Gnai2 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P59215 | Gnao1 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P08753 | Gnai3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| Q6Q7Y5 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Rattus norvegicus (Rat) | SS |
| Q0DJ33 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Oryza sativa subsp. japonica (Rice) | SS |
| P51875 | goa-1 | Guanine nucleotide-binding protein G | Caenorhabditis elegans | SS |
| Q18434 | odr-3 | Guanine nucleotide-binding protein alpha-17 subunit | Caenorhabditis elegans | SS |
| Q9BIG4 | gpa-10 | Guanine nucleotide-binding protein alpha-10 subunit | Caenorhabditis elegans | PR |
| P28052 | gpa-3 | Guanine nucleotide-binding protein alpha-3 subunit | Caenorhabditis elegans | SS |
| Q9XTB2 | gpa-13 | Guanine nucleotide-binding protein alpha-13 subunit | Caenorhabditis elegans | SS |
| Q9BIG5 | gpa-4 | Guanine nucleotide-binding protein alpha-4 subunit | Caenorhabditis elegans | SS |
| P28051 | gpa-1 | Guanine nucleotide-binding protein alpha-1 subunit | Caenorhabditis elegans | SS |
| Q9N2V6 | gpa-16 | Guanine nucleotide-binding protein alpha-16 subunit | Caenorhabditis elegans | SS |
| P91907 | gpa-15 | Guanine nucleotide-binding protein alpha-15 subunit | Caenorhabditis elegans | SS |
| Q20907 | gpa-8 | Guanine nucleotide-binding protein alpha-8 subunit | Caenorhabditis elegans | PR |
| Q21917 | gpa-7 | Guanine nucleotide-binding protein alpha-7 subunit | Caenorhabditis elegans | SS |
| Q93743 | gpa-6 | Guanine nucleotide-binding protein alpha-6 subunit | Caenorhabditis elegans | SS |
| O76584 | gpa-11 | Guanine nucleotide-binding protein alpha-11 subunit | Caenorhabditis elegans | SS |
| P49084 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| P93163 | GPA2 | Guanine nucleotide-binding protein alpha-2 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| O80462 | XLG1 | Extra-large guanine nucleotide-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P18064 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P26981 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGAGASAEEK | HSRELEKKLK | EDAEKDARTV | KLLLLGAGES | GKSTIVKQMK | IIHQDGYSLE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ECLEFIAIIY | GNTLQSILAI | VRAMTTLNIQ | YGDSARQDDA | RKLMHMADTI | EEGTMPKEMS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DIIQRLWKDS | GIQACFERAS | EYQLNDSAGY | YLSDLERLVT | PGYVPTEQDV | LRSRVKTTGI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IETQFSFKDL | NFRMFDVGGQ | RSERKKWIHC | FEGVTCIIFI | AALSAYDMVL | VEDDEVNRMH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ESLHLFNSIC | NHRYFATTSI | VLFLNKKDVF | FEKIKKAHLS | ICFPDYDGPN | TYEDAGNYIK |
| 310 | 320 | 330 | 340 | ||
| VQFLELNMRR | DVKEIYSHMT | CATDTQNVKF | VFDAVTDIII | KENLKDCGLF |