P10644
Gene name |
PRKAR1A (PKR1, PRKAR1, TSE1) |
Protein name |
cAMP-dependent protein kinase type I-alpha regulatory subunit |
Names |
Tissue-specific extinguisher 1, TSE1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5573 |
EC number |
|
Protein Class |
CAMP-DEPENDENT PROTEIN KINASE REGULATORY CHAIN (PTHR11635) |
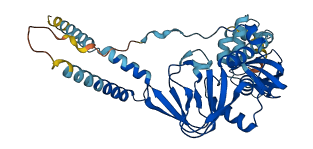
Descriptions
PKA is a major intracellular receptor of cAMP and modulates diverse cellular signalings. The N-terminal regulatory domain contains a pseudosubstrate sequence that may interact with the catalytic kinase domain.
Autoinhibitory domains (AIDs)
Target domain |
137-253 (Cyclic nucleotide-binding domain) |
Relief mechanism |
Ligand binding |
Assay |
Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for P10644
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 5KJX | X-ray | 190 A | A | 234-381 | PDB |
| 5KJY | X-ray | 200 A | A | 234-381 | PDB |
| 5KJZ | X-ray | 135 A | A | 234-381 | PDB |
| AF-P10644-F1 | Predicted | AlphaFoldDB |
233 variants for P10644
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs281864779 RCV000013505 RCV000523178 |
1 | M>V | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1600461789 CA400751780 RCV001011392 |
5 | S>G | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs369210646 RCV000701277 CA8729138 RCV000562330 |
7 | A>T | Hereditary cancer-predisposing syndrome Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs745391692 RCV001016643 CA8729142 RCV001856199 RCV000781768 |
9 | S>R | Hereditary cancer-predisposing syndrome Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000265264 RCV000558438 rs886041228 CA10603402 |
16 | R>* | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP |
|
RCV001024779 rs1600461989 CA400751882 |
20 | L>R | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA10583669 RCV000232417 rs878854561 |
22 | V>F | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000466415 CA8729151 rs763158372 |
24 | K>R | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA400751929 rs1235317386 RCV001853793 RCV000575969 |
27 | I>M | Carney complex, type 1 Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
rs281864780 RCV000034296 CA344454 |
28 | Q>* | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001017705 rs1600462191 CA400751932 |
28 | Q>P | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000034298 rs281864787 |
29 | A>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs760726941 RCV000553000 CA8729154 |
29 | A>S | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
RCV000556130 rs756985434 CA8729157 |
32 | K>E | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs281864788 RCV000034282 |
34 | S>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000704942 CA293327597 rs944887425 |
34 | S>C | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
RCV000323950 CA8729158 rs377513504 RCV000541002 |
35 | I>V | Acrodysostosis 1 with or without hormone resistance Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1600462346 RCV001009840 CA400751983 |
36 | V>G | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA344417 RCV000034283 rs281864781 |
37 | Q>* | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA400752011 rs1356757839 RCV000645597 |
41 | A>T | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000034284 COSM1479905 CA344422 rs281864782 |
42 | R>* | Carney complex, type 1 breast [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar ExAC dbSNP gnomAD |
|
rs281864789 RCV000034285 |
47 | M>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA293327658 RCV000695293 rs967277610 |
51 | R>G | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP |
|
rs2085656304 RCV001253023 |
60 | E>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000813553 rs769706055 CA8729194 |
73 | T>S | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001281678 rs2085657588 |
74 | R>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA256533 RCV000148738 VAR_046895 RCV000013510 rs137853303 |
74 | R>C | CNC1; exhibits increased PKA activity which is attributed to decreased binding to cAMP and/or the catalytic subunit Carney complex, type 1 Carney complex [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt ESP TOPMed dbSNP gnomAD |
|
RCV000561393 rs200069356 CA8729195 RCV000384218 RCV000588205 |
74 | R>H | Acrodysostosis 1 with or without hormone resistance Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs1600478702 CA400752282 RCV001015311 |
79 | E>G | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1600478753 RCV001015769 CA400752316 |
84 | P>S | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA8729199 rs759197325 RCV000556071 |
91 | V>I | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
rs281864783 RCV000034288 CA344434 |
96 | R>* | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA400752390 rs1555813215 RCV000645593 |
96 | R>Q | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000691927 CA400752409 rs1568695224 |
99 | A>V | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000505608 COSM148322 rs1194755479 RCV000544572 CA400752486 |
110 | A>V | Medulloblastoma Carney complex, type 1 stomach [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar TOPMed dbSNP gnomAD |
|
RCV000703002 rs1472324247 CA400752487 |
111 | A>T | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
rs2085693506 RCV001038140 |
144 | N>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA400752830 RCV000704084 rs1568696484 |
155 | S>L | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA8729265 rs776071358 RCV000536758 |
157 | S>F | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
rs765675434 CA8729267 RCV000645603 |
160 | A>T | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000414608 RCV000013498 rs281864790 |
164 | V>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001223560 rs2085739247 |
178 | D>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000429583 rs141913727 RCV000509168 CA16607826 |
179 | Q>* | Carney complex [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA8729298 rs199801675 RCV000493890 RCV000692010 |
182 | T>M | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000034291 rs281864792 |
189 | E>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA8729321 RCV000534903 RCV000587029 rs767405408 |
189 | E>D | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000645591 CA400753098 rs1555814054 |
191 | A>T | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA8729323 RCV000528487 rs755798109 |
199 | S>N | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000013501 rs281864791 |
206 | I>missing | Familial atrial myxoma [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1600486197 CA400753208 RCV000791348 |
207 | Y>C | Acrodysostosis 1 with or without hormone resistance [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs727503379 RCV000317950 RCV000151681 |
208 | G>missing | Carney complex [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000034292 CA344442 VAR_046898 COSM26705 rs281864786 |
213 | A>D | CNC1; exhibits increased PKA activity which is attributed to decreased binding to cAMP and/or the catalytic subunit; reduces protein degradation Carney complex, type 1 soft_tissue [UniProt, ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar UniProt Ensembl dbSNP |
|
rs1555814093 RCV001025310 RCV000588769 CA400753263 |
216 | K>R | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000691594 rs1568698487 |
221 | V>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000697266 rs1568698504 CA400753317 |
224 | W>* | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs281864784 RCV000627207 CA344447 RCV000034293 |
228 | R>* | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs281864793 RCV000034294 |
232 | R>* | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs281864794 RCV000034295 |
238 | S>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1555814475 RCV000539006 CA400753585 |
254 | S>C | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1555814477 RCV000645601 |
255 | I>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000542202 rs1255519868 CA400753794 |
260 | D>H | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP |
|
RCV000688438 rs1568701297 CA400753805 |
261 | K>R | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000013499 rs281864785 CA341220 |
262 | W>C* | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001026953 RCV001194345 CA400753828 rs1303569195 |
264 | R>H | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000696479 rs1568701362 |
271 | L>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1600492296 CA400753930 RCV001017641 |
279 | G>E | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1568701408 RCV000690842 CA400753934 |
280 | Q>E | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000034297 rs281864795 |
283 | V>missing | Carney complex, type 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1555814719 RCV000595124 RCV001854100 VAR_069459 CA400753971 |
285 | Q>R | ACRDYS1; reduces PKA activity; decreases cAMP binding Carney complex, type 1 [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA8729414 rs768253469 RCV000564440 |
305 | R>H | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA400754201 rs1600494933 RCV001019244 |
313 | V>F | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs387906695 VAR_069461 RCV000022794 CA128754 |
327 | I>T | Acrodysostosis 1 with or without hormone resistance ACRDYS1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA400754491 rs1555815121 RCV000763414 RCV000497832 |
335 | R>C | Acrodysostosis 1 with or without hormone resistance Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
VAR_069463 rs387906694 CA128749 RCV000022793 |
335 | R>P | Acrodysostosis 1 with or without hormone resistance ACRDYS1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs146383819 CA8729456 RCV000459769 |
342 | R>C | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs760033566 RCV000645594 CA8729457 |
342 | R>H | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
CA8729459 RCV000459182 rs772571340 |
348 | V>I | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA400754788 RCV000801888 rs1600496410 |
360 | G>V | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000645595 CA400754821 rs1555815165 |
363 | S>L | Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000022791 RCV001824573 RCV000760318 CA128739 rs387906692 RCV001852002 |
368 | R>* | Pigmented nodular adrenocortical disease, primary, 1 Acrodysostosis 1 with or without hormone resistance Carney complex, type 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
VAR_068241 RCV000022792 rs387906693 CA128744 |
373 | Y>H | Acrodysostosis 1 with or without hormone resistance ACRDYS1 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1568703465 RCV001009960 RCV000780649 CA400755012 |
378 | S>L | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA400751761 rs1600461762 |
2 | E>Q | No |
ClinGen Ensembl |
|
|
rs752643409 CA8729136 |
3 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA400751769 rs1488444958 |
3 | S>P | No |
ClinGen TOPMed |
|
|
CA400751779 rs1600461789 |
5 | S>R | No |
ClinGen Ensembl |
|
|
rs1555811666 RCV000479092 |
6 | T>missing | No |
ClinVar dbSNP |
|
|
CA400751787 rs1600461810 |
6 | T>P | No |
ClinGen Ensembl |
|
|
rs1224297486 CA400751798 |
8 | A>T | No |
ClinGen TOPMed |
|
|
rs778468626 CA400751805 |
9 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA8729141 rs778468626 |
9 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs1467886544 CA400751810 |
10 | E>K | No |
ClinGen gnomAD |
|
|
rs1292133703 CA400751830 |
12 | A>G | No |
ClinGen TOPMed |
|
|
rs1177674637 CA400751832 |
13 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1177674637 CA400751834 |
13 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs757185291 CA293327486 |
14 | S>N | No |
ClinGen gnomAD |
|
|
CA400751841 rs757185291 |
14 | S>T | No |
ClinGen gnomAD |
|
|
CA8729143 rs771518581 |
17 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400751865 rs1172984891 |
18 | C>Y | No |
ClinGen Ensembl |
|
|
rs780569077 CA8729144 |
19 | E>A | No |
ClinGen ExAC gnomAD |
|
|
rs769019433 CA8729146 |
21 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs770156593 CA8729149 |
23 | Q>R | No |
ClinGen ExAC |
|
|
RCV000481932 rs1064793709 |
24 | K>missing | No |
ClinVar dbSNP |
|
|
CA8729152 rs140795787 |
25 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8729153 rs774277428 |
26 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8729155 rs763929951 |
29 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400751941 rs1202375076 |
30 | L>V | No |
ClinGen gnomAD |
|
|
rs1176097601 CA400751955 |
32 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA400751969 rs1257543736 |
34 | S>A | No |
ClinGen gnomAD |
|
|
CA400751980 rs1215353269 |
36 | V>L | No |
ClinGen TOPMed |
|
|
CA8729160 rs758013363 |
40 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA400752016 rs1460961845 |
41 | A>D | No |
ClinGen TOPMed gnomAD |
|
|
CA400752015 rs1460961845 |
41 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA8729161 rs281864782 |
42 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA8729162 rs746486900 |
42 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
RCV000478061 rs1555811735 |
43 | P>missing | No |
ClinVar dbSNP |
|
|
rs1568688352 CA400752021 |
43 | P>S | No |
ClinGen Ensembl |
|
|
rs548529083 CA293327657 |
47 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA400752070 rs1214905938 |
50 | L>F | No |
ClinGen gnomAD |
|
|
CA8729165 rs748547646 |
52 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA400752111 rs1471017649 |
56 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA8729167 rs773684158 |
56 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs772001678 CA8729191 |
61 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA8729192 rs775514809 |
62 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1438564398 CA400752186 |
64 | Q>H | No |
ClinGen gnomAD |
|
|
CA400752196 rs1600478559 |
66 | Q>K | No |
ClinGen Ensembl |
|
|
CA293288905 rs920361727 |
69 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA400752234 rs1281684412 |
71 | A>V | No |
ClinGen gnomAD |
|
|
RCV000481239 rs1064793712 |
72 | G>missing | No |
ClinVar dbSNP |
|
|
rs769706055 CA400752245 |
73 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400752249 rs200069356 |
74 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA8729196 rs762678902 |
80 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1470545069 CA400752303 |
82 | I>F | No |
ClinGen gnomAD |
|
|
rs267605013 CA293288926 |
85 | P>L | No |
ClinGen Ensembl |
|
|
CA293288919 rs267605012 |
85 | P>S | No |
ClinGen TOPMed |
|
|
CA400752321 rs267605012 |
85 | P>T | No |
ClinGen TOPMed |
|
|
CA400752333 rs1165953688 |
87 | P>L | No |
ClinGen gnomAD |
|
|
rs1456959022 CA400752332 |
87 | P>S | No |
ClinGen gnomAD |
|
|
CA400752340 rs1396360533 |
88 | N>S | No |
ClinGen gnomAD |
|
|
CA400752345 rs1459684511 |
89 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1459684511 CA400752346 |
89 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1443738914 CA400752375 |
94 | R>G | No |
ClinGen gnomAD |
|
|
CA400752394 rs1555813217 RCV000627206 |
97 | R>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs779224934 CA8729206 |
103 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1568695288 CA400752440 |
104 | V>I | No |
ClinGen Ensembl |
|
|
CA293288957 rs748583171 |
105 | Y>H | No |
ClinGen Ensembl |
|
|
CA400752455 rs1600479149 COSM1385477 |
106 | T>M | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA400752460 rs1600479179 |
107 | E>A | No |
ClinGen Ensembl |
|
|
rs1206688715 CA400752477 |
109 | D>G | No |
ClinGen gnomAD |
|
|
CA400752489 rs1472324247 |
111 | A>S | No |
ClinGen gnomAD |
|
|
rs1310506395 CA400752497 |
112 | S>F | No |
ClinGen TOPMed |
|
|
CA293288984 rs267605014 |
115 | R>K | No |
ClinGen Ensembl |
|
|
CA400752551 rs1253034687 |
119 | P>T | No |
ClinGen TOPMed |
|
|
RCV000369351 rs886041530 |
121 | D>missing | No |
ClinVar dbSNP |
|
|
CA400752591 rs1205068188 |
124 | T>A | No |
ClinGen TOPMed |
|
|
rs886041457 RCV000371626 |
125 | M>missing | No |
ClinVar dbSNP |
|
|
rs1555813434 RCV000483450 |
129 | A>missing | No |
ClinVar dbSNP |
|
|
rs1383865866 CA400752665 |
134 | K>N | No |
ClinGen TOPMed |
|
|
rs1064794280 RCV000483496 |
136 | V>missing | No |
ClinVar dbSNP |
|
|
rs781219513 CA8729233 |
141 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA400752723 rs1296031514 |
143 | D>G | No |
ClinGen gnomAD |
|
|
CA8729234 rs749233975 |
143 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA400752769 rs1296802743 |
147 | S>R | No |
ClinGen TOPMed |
|
|
CA400752771 rs1384256151 |
148 | D>Y | No |
ClinGen TOPMed |
|
|
CA8729264 rs768408952 |
156 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1269379592 CA400752846 |
158 | F>C | No |
ClinGen gnomAD |
|
|
CA400752855 rs372389205 |
159 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400752857 rs765675434 |
160 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1064797015 RCV000479640 |
163 | T>missing | No |
ClinVar dbSNP |
|
|
CA293289854 rs867753055 |
166 | Q>* | No |
ClinGen Ensembl |
|
|
CA400752932 rs1600484143 |
169 | D>G | No |
ClinGen Ensembl |
|
|
rs148109898 CA400752973 |
174 | F>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
RCV001008377 rs1600484162 |
175 | Y>missing | No |
ClinVar dbSNP |
|
|
RCV000121881 rs587778626 CA161650 |
177 | I>T | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs141913727 CA8729296 |
179 | Q>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA293291297 rs773162575 |
184 | V>I | No |
ClinGen Ensembl |
|
|
rs369011860 CA8729320 |
185 | Y>C | No |
ClinGen ESP ExAC gnomAD |
|
|
CA400753088 rs767405408 |
189 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752432156 CA8729322 |
191 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400753110 rs1600486090 |
193 | S>G | No |
ClinGen Ensembl |
|
|
rs1600486256 CA400753223 |
209 | T>I | No |
ClinGen Ensembl |
|
|
rs917497011 CA293291349 |
210 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA8729328 rs746418827 |
212 | A>P | No |
ClinGen ExAC gnomAD |
|
|
rs950375633 CA293291356 |
218 | K>E | No |
ClinGen TOPMed |
|
|
rs1044570185 CA293291364 |
223 | L>W | No |
ClinGen TOPMed |
|
|
rs1555814100 RCV000519219 CA400753319 |
224 | W>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA8729332 rs768934933 |
228 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA8729333 rs768934933 |
228 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA8729334 rs762014574 |
229 | D>N | No |
ClinGen ExAC gnomAD |
|
|
RCV001092734 rs2085775108 |
232 | R>K | No |
ClinVar dbSNP |
|
|
rs1555814115 RCV000519355 |
233 | R>missing | No |
ClinVar dbSNP |
|
|
CA400753443 rs1483410726 |
241 | R>T | No |
ClinGen gnomAD |
|
|
rs879148650 CA293292716 |
242 | K>T | No |
ClinGen Ensembl |
|
|
CA8729356 rs762880866 |
245 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs773430995 CA8729355 |
245 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
RCV000349156 rs886041351 CA10603600 |
246 | Y>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs376701965 CA8729357 |
247 | E>D | No |
ClinGen ESP ExAC gnomAD |
|
|
rs775247484 CA8729358 |
250 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA8729360 rs763921895 |
252 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1242606073 CA400753566 |
252 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1431068317 COSM267259 CA400753574 |
253 | V>A | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA400753586 rs1282833520 |
255 | I>V | No |
ClinGen TOPMed |
|
|
CA8729391 rs755544940 |
261 | K>E | No |
ClinGen ExAC |
|
|
rs1329426125 CA400753842 |
266 | T>M | No |
ClinGen gnomAD |
|
|
rs1085307672 RCV000489158 CA400753859 |
269 | D>G | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA8729394 rs756503981 |
270 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA400753915 rs1320674630 |
277 | E>G | No |
ClinGen TOPMed |
|
|
rs1383606243 CA400753945 |
281 | K>R | No |
ClinGen TOPMed |
|
|
rs749518806 CA8729396 |
282 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1264572252 CA400753963 |
284 | V>L | No |
ClinGen gnomAD |
|
|
rs1264572252 CA400753961 |
284 | V>M | No |
ClinGen gnomAD |
|
|
rs1413778575 CA400753990 |
288 | P>A | No |
ClinGen gnomAD |
|
|
CA8729411 rs765989838 |
300 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA400754107 rs1424094903 |
303 | L>R | No |
ClinGen gnomAD |
|
|
rs1568702564 RCV000781767 CA400754123 |
306 | R>Q | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA8729415 rs753294119 |
308 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs756663310 CA8729416 |
309 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA8729417 rs764420601 |
310 | E>A | No |
ClinGen ExAC gnomAD |
|
|
CA400754214 rs1199251597 |
314 | E>G | No |
ClinGen gnomAD |
|
|
rs1057517805 RCV000413468 |
317 | R>missing | No |
ClinVar dbSNP |
|
|
CA400754269 rs1156315838 |
319 | G>E | No |
ClinGen TOPMed |
|
|
rs1383730251 CA400754266 |
319 | G>R | No |
ClinGen TOPMed |
|
|
CA8729420 rs779141257 |
320 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1399973055 CA400754539 |
339 | V>A | No |
ClinGen gnomAD |
|
|
CA400754593 rs1600496295 |
344 | P>L | No |
ClinGen Ensembl |
|
|
CA400754617 rs1466357740 |
346 | K>R | No |
ClinGen TOPMed |
|
|
rs1473853442 CA400754729 |
355 | F>C | No |
ClinGen TOPMed |
|
|
CA400754796 rs1600496423 |
361 | P>A | No |
ClinGen Ensembl |
|
|
rs751660273 CA8729464 COSM437218 |
364 | D>Y | breast [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA293294615 rs201332278 |
369 | N>D | No |
ClinGen 1000Genomes |
|
|
rs1268436057 CA400754923 |
372 | Q>R | No |
ClinGen gnomAD |
|
|
RCV000414079 rs1057517806 |
374 | N>missing | No |
ClinVar dbSNP |
|
|
rs1184620103 CA400754960 |
374 | N>S | No |
ClinGen gnomAD |
4 associated diseases with P10644
[MIM: 160980]: Carney complex 1 (CNC1)
CNC is a multiple neoplasia syndrome characterized by spotty skin pigmentation, cardiac and other myxomas, endocrine tumors, and psammomatous melanotic schwannomas. {ECO:0000269|PubMed:15371594, ECO:0000269|PubMed:18241045, ECO:0000269|PubMed:22785148, ECO:0000269|PubMed:23323113, ECO:0000269|PubMed:26405036}. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 255960]: Intracardiac myxoma (INTMYX)
Inheritance is autosomal recessive. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 610489]: Primary pigmented nodular adrenocortical disease 1 (PPNAD1)
A rare bilateral adrenal defect causing ACTH-independent Cushing syndrome. Macroscopic appearance of the adrenals is characteristic with small pigmented micronodules observed in the cortex. Clinical manifestations of Cushing syndrome include facial and truncal obesity, abdominal striae, muscular weakness, osteoporosis, arterial hypertension, diabetes. PPNAD1 is most often diagnosed in patients with Carney complex, a multiple neoplasia syndrome. However it can also be observed in patients without other manifestations. {ECO:0000269|PubMed:12213893}. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 101800]: Acrodysostosis 1, with or without hormone resistance (ACRDYS1)
A form of skeletal dysplasia characterized by short stature, severe brachydactyly, facial dysostosis, and nasal hypoplasia. Affected individuals often have advanced bone age and obesity. Laboratory studies show resistance to multiple hormones, including parathyroid, thyrotropin, calcitonin, growth hormone-releasing hormone, and gonadotropin. However, not all patients show endocrine abnormalities. {ECO:0000269|PubMed:21651393, ECO:0000269|PubMed:22464250, ECO:0000269|PubMed:22464252, ECO:0000269|PubMed:22723333, ECO:0000269|PubMed:23043190, ECO:0000269|PubMed:23425300, ECO:0000269|PubMed:26405036}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- CNC is a multiple neoplasia syndrome characterized by spotty skin pigmentation, cardiac and other myxomas, endocrine tumors, and psammomatous melanotic schwannomas. {ECO:0000269|PubMed:15371594, ECO:0000269|PubMed:18241045, ECO:0000269|PubMed:22785148, ECO:0000269|PubMed:23323113, ECO:0000269|PubMed:26405036}. Note=The disease is caused by variants affecting the gene represented in this entry.
- Inheritance is autosomal recessive. Note=The disease is caused by variants affecting the gene represented in this entry.
- A rare bilateral adrenal defect causing ACTH-independent Cushing syndrome. Macroscopic appearance of the adrenals is characteristic with small pigmented micronodules observed in the cortex. Clinical manifestations of Cushing syndrome include facial and truncal obesity, abdominal striae, muscular weakness, osteoporosis, arterial hypertension, diabetes. PPNAD1 is most often diagnosed in patients with Carney complex, a multiple neoplasia syndrome. However it can also be observed in patients without other manifestations. {ECO:0000269|PubMed:12213893}. Note=The disease is caused by variants affecting the gene represented in this entry.
- A form of skeletal dysplasia characterized by short stature, severe brachydactyly, facial dysostosis, and nasal hypoplasia. Affected individuals often have advanced bone age and obesity. Laboratory studies show resistance to multiple hormones, including parathyroid, thyrotropin, calcitonin, growth hormone-releasing hormone, and gonadotropin. However, not all patients show endocrine abnormalities. {ECO:0000269|PubMed:21651393, ECO:0000269|PubMed:22464250, ECO:0000269|PubMed:22464252, ECO:0000269|PubMed:22723333, ECO:0000269|PubMed:23043190, ECO:0000269|PubMed:23425300, ECO:0000269|PubMed:26405036}. Note=The disease is caused by variants affecting the gene represented in this entry.
7 regional properties for P10644
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Cyclic nucleotide-binding domain | 137 - 253 | IPR000595-1 |
| domain | Cyclic nucleotide-binding domain | 255 - 376 | IPR000595-2 |
| domain | cAMP-dependent protein kinase regulatory subunit, dimerization-anchoring domain | 25 - 62 | IPR003117 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 164 - 180 | IPR018488-1 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 200 - 217 | IPR018488-2 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 282 - 298 | IPR018488-3 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 324 - 341 | IPR018488-4 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11635 | CAMP-DEPENDENT PROTEIN KINASE REGULATORY CHAIN |
| PANTHER Subfamily | PTHR11635:SF129 | CAMP-DEPENDENT PROTEIN KINASE TYPE I-ALPHA REGULATORY SUBUNIT |
| PANTHER Protein Class | kinase modulator | |
| PANTHER Pathway Category |
Metabotropic glutamate receptor group III pathway PKA Muscarinic acetylcholine receptor 2 and 4 signaling pathway PKA GABA-B receptor II signaling PKA Transcription regulation by bZIP transcription factor PKA Metabotropic glutamate receptor group II pathway PKA Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway PKA-r Hedgehog signaling pathway PKA Endothelin signaling pathway PKA Metabotropic glutamate receptor group I pathway PKA |
|
13 GO annotations of cellular component
| Name | Definition |
|---|---|
| cAMP-dependent protein kinase complex | An enzyme complex, composed of regulatory and catalytic subunits, that catalyzes protein phosphorylation. Inactive forms of the enzyme have two regulatory chains and two catalytic chains; activation by cAMP produces two active catalytic monomers and a regulatory dimer. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| ciliary base | Area of the cilium (also called flagellum) where the basal body and the axoneme are anchored to the plasma membrane. The ciliary base encompasses the distal part of the basal body, transition fibers and transition zone and is structurally and functionally very distinct from the rest of the cilium. In this area proteins are sorted and filtered before entering the cilium, and many ciliary proteins localize specifically to this area. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| multivesicular body | A type of endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| neuromuscular junction | The junction between the axon of a motor neuron and a muscle fiber. In response to the arrival of action potentials, the presynaptic button releases molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane of the muscle fiber, leading to a change in post-synaptic potential. |
| nucleotide-activated protein kinase complex | A protein complex that possesses nucleotide-dependent protein kinase activity. The nucleotide can be AMP (in S. pombe and human) or ADP (in S. cerevisiae). |
| plasma membrane raft | A membrane raft that is part of the plasma membrane. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| sperm connecting piece | The segment of the sperm flagellum that attaches to the implantation fossa of the nucleus in the sperm head; from the remnant of the centriole at this point, the axoneme extends throughout the length of the flagellum. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| cAMP binding | Binding to cAMP, the nucleotide cyclic AMP (adenosine 3',5'-cyclophosphate). |
| cAMP-dependent protein kinase inhibitor activity | Binds to and stops, prevents or reduces the activity of a cAMP-dependent protein kinase. |
| cAMP-dependent protein kinase regulator activity | Modulation of the activity of the enzyme cAMP-dependent protein kinase. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase A catalytic subunit binding | Binding to one or both of the catalytic subunits of protein kinase A. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| cardiac muscle cell proliferation | The expansion of a cardiac muscle cell population by cell division. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| mesoderm formation | The process that gives rise to the mesoderm. This process pertains to the initial formation of the structure from unspecified parts. |
| negative regulation of activated T cell proliferation | Any process that stops, prevents or reduces the rate or extent of activated T cell proliferation. |
| negative regulation of cAMP-dependent protein kinase activity | Any process that stops, prevents or reduces the frequency, rate or extent of cAMP-dependent protein kinase activity. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| sarcomere organization | The myofibril assembly process that results in the organization of muscle actomyosin into sarcomeres. The sarcomere is the repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs. |
22 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P31322 | PRKAR2B | cAMP-dependent protein kinase type II-beta regulatory subunit | Bos taurus (Bovine) | EV |
| P00515 | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit | Bos taurus (Bovine) | EV |
| P00514 | PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | Bos taurus (Bovine) | EV |
| Q5ZM91 | PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | Gallus gallus (Chicken) | SS |
| P81900 | Pka-R2 | cAMP-dependent protein kinase type II regulatory subunit | Drosophila melanogaster (Fruit fly) | PR |
| P16905 | Pka-R1 | cAMP-dependent protein kinase type I regulatory subunit | Drosophila melanogaster (Fruit fly) | PR |
| Q96M20 | CNBD2 | Cyclic nucleotide-binding domain-containing protein 2 | Homo sapiens (Human) | PR |
| P13861 | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit | Homo sapiens (Human) | PR |
| P31323 | PRKAR2B | cAMP-dependent protein kinase type II-beta regulatory subunit | Homo sapiens (Human) | PR |
| P31321 | PRKAR1B | cAMP-dependent protein kinase type I-beta regulatory subunit | Homo sapiens (Human) | EV |
| P12367 | Prkar2a | cAMP-dependent protein kinase type II-alpha regulatory subunit | Mus musculus (Mouse) | PR |
| Q9D5U8 | Cnbd2 | Cyclic nucleotide-binding domain-containing protein 2 | Mus musculus (Mouse) | PR |
| P12849 | Prkar1b | cAMP-dependent protein kinase type I-beta regulatory subunit | Mus musculus (Mouse) | PR |
| P31324 | Prkar2b | cAMP-dependent protein kinase type II-beta regulatory subunit | Mus musculus (Mouse) | PR |
| Q9DBC7 | Prkar1a | cAMP-dependent protein kinase type I-alpha regulatory subunit [Cleaved into: cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed] | Mus musculus (Mouse) | SS |
| P05207 | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit | Sus scrofa (Pig) | SS |
| P07802 | PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | Sus scrofa (Pig) | SS |
| P81377 | Prkar1b | cAMP-dependent protein kinase type I-beta regulatory subunit | Rattus norvegicus (Rat) | PR |
| P12368 | Prkar2a | cAMP-dependent protein kinase type II-alpha regulatory subunit | Rattus norvegicus (Rat) | PR |
| P12369 | Prkar2b | cAMP-dependent protein kinase type II-beta regulatory subunit | Rattus norvegicus (Rat) | PR |
| P09456 | Prkar1a | cAMP-dependent protein kinase type I-alpha regulatory subunit | Rattus norvegicus (Rat) | SS |
| P30625 | kin-2 | cAMP-dependent protein kinase regulatory subunit | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MESGSTAASE | EARSLRECEL | YVQKHNIQAL | LKDSIVQLCT | ARPERPMAFL | REYFERLEKE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EAKQIQNLQK | AGTRTDSRED | EISPPPPNPV | VKGRRRRGAI | SAEVYTEEDA | ASYVRKVIPK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DYKTMAALAK | AIEKNVLFSH | LDDNERSDIF | DAMFSVSFIA | GETVIQQGDE | GDNFYVIDQG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ETDVYVNNEW | ATSVGEGGSF | GELALIYGTP | RAATVKAKTN | VKLWGIDRDS | YRRILMGSTL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RKRKMYEEFL | SKVSILESLD | KWERLTVADA | LEPVQFEDGQ | KIVVQGEPGD | EFFIILEGSA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| AVLQRRSENE | EFVEVGRLGP | SDYFGEIALL | MNRPRAATVV | ARGPLKCVKL | DRPRFERVLG |
| 370 | 380 | ||||
| PCSDILKRNI | QQYNSFVSLS | V |