P0CW00
Gene name |
TSPY8 |
Protein name |
Testis-specific Y-encoded protein 8 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:728403 |
EC number |
|
Protein Class |
|
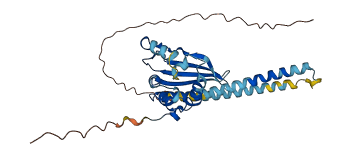
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P0CW00
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P0CW00-F1 | Predicted | AlphaFoldDB |
46 variants for P0CW00
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1603140079 CA414787533 |
2 | R>C | No |
ClinGen Ensembl |
|
|
rs760572223 CA10572407 |
3 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs753631588 CA10572409 |
5 | G>C | No |
ClinGen ExAC gnomAD |
|
|
CA10572410 rs758572301 |
5 | G>D | No |
ClinGen ExAC |
|
|
rs757408537 CA10572413 |
7 | L>M | No |
ClinGen ExAC |
|
|
rs780310523 CA414787650 CA10572417 |
10 | W>C | No |
ClinGen ExAC gnomAD |
|
|
CA10572418 rs749613655 |
11 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA10572419 rs1556180031 |
13 | E>G | No |
ClinGen Ensembl |
|
|
CA10572421 rs768306591 |
14 | R>S | No |
ClinGen ExAC gnomAD |
|
|
rs1476646611 CA414787750 |
18 | G>C | No |
ClinGen gnomAD |
|
|
rs747718289 CA10572423 |
20 | C>Y | No |
ClinGen ExAC |
|
|
CA414787798 rs1172231287 |
22 | V>M | No |
ClinGen gnomAD |
|
|
CA10572424 rs771553199 |
23 | G>S | No |
ClinGen ExAC |
|
|
rs773243424 CA414787822 |
24 | R>W | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 39 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA414788157 rs1464787539 |
48 | G>R | No |
ClinGen gnomAD |
|
|
rs1464787539 CA414788158 |
48 | G>W | No |
ClinGen gnomAD |
|
| TCGA novel | 57 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA414788348 rs1603140150 |
62 | E>* | No |
ClinGen Ensembl |
|
| TCGA novel | 77 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA414789284 rs1556180134 |
131 | S>T | No |
ClinGen Ensembl |
|
|
CA414789477 rs1328941043 |
144 | H>Q | No |
ClinGen gnomAD |
|
|
CA414789509 rs1556180155 |
147 | R>C | No |
ClinGen Ensembl |
|
|
rs1432728779 CA414791239 |
232 | S>Y | No |
ClinGen gnomAD |
|
|
rs1603140212 CA414791291 |
235 | I>S | No |
ClinGen Ensembl |
|
|
rs1278931930 CA414791280 |
235 | I>V | No |
ClinGen gnomAD |
|
|
CA10572426 rs760563458 |
237 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA414791348 rs760563458 |
237 | W>C | No |
ClinGen ExAC gnomAD |
|
|
rs1216872203 CA645293768 |
238 | Y>* | No |
ClinGen gnomAD |
|
|
rs410975 CA337637650 |
239 | L>P | No |
ClinGen Ensembl |
|
|
CA414791416 rs1603140251 |
240 | D>E | No |
ClinGen Ensembl |
|
|
CA414791410 rs1603140245 |
240 | D>G | No |
ClinGen Ensembl |
|
|
CA414791399 rs1603140240 |
240 | D>H | No |
ClinGen Ensembl |
|
|
CA414791447 rs1603140259 |
242 | E>* | No |
ClinGen Ensembl |
|
|
rs1238925689 CA414791508 |
244 | E>G | No |
ClinGen Ensembl |
|
|
rs1603140269 CA414791553 |
246 | Y>* | No |
ClinGen Ensembl |
|
|
CA10572427 rs770855086 |
246 | Y>D | No |
ClinGen ExAC |
|
|
CA10572428 rs78649013 |
247 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA414791558 rs78649013 |
247 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA10572429 rs759309586 |
248 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA10572430 rs764478973 |
259 | N>H | No |
ClinGen ExAC gnomAD |
|
|
CA414791892 rs1477246214 |
263 | D>N | No |
ClinGen gnomAD |
|
|
rs1556180322 CA414791995 |
269 | S>F | No |
ClinGen Ensembl |
|
|
COSM4157084 rs368632690 CA10572431 |
304 | S>F | thyroid [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA414792681 rs1603140324 |
305 | Q>H | No |
ClinGen Ensembl |
|
|
CA414792738 rs1603140329 |
309 | S>S | No |
ClinGen Ensembl |
No associated diseases with P0CW00
No regional properties for P0CW00
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P0CW00 | |||
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| histone binding | Binding to a histone, any of a group of water-soluble proteins found in association with the DNA of eukaryotic or archaeal chromosomes. They are involved in the condensation and coiling of chromosomes during cell division and have also been implicated in gene regulation and DNA replication. They may be chemically modified (methylated, acetlyated and others) to regulate gene transcription. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| gonadal mesoderm development | The process whose specific outcome is the progression of the gonadal mesoderm over time, from its formation to the mature structure. The gonadal mesoderm is the middle layer of the three primary germ layers of the embryo which will go on to form the gonads of the organism. |
| nucleosome assembly | The aggregation, arrangement and bonding together of a nucleosome, the beadlike structural units of eukaryotic chromatin composed of histones and DNA. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O19110 | TSPY1 | Testis-specific Y-encoded protein 1 | Bos taurus (Bovine) | PR |
| Q99457 | NAP1L3 | Nucleosome assembly protein 1-like 3 | Homo sapiens (Human) | PR |
| P55209 | NAP1L1 | Nucleosome assembly protein 1-like 1 | Homo sapiens (Human) | PR |
| Q01534 | TSPY1 | Testis-specific Y-encoded protein 1 | Homo sapiens (Human) | PR |
| P0CV99 | TSPY4 | Testis-specific Y-encoded protein 4 | Homo sapiens (Human) | PR |
| P0CV98 | TSPY3 | Testis-specific Y-encoded protein 3 | Homo sapiens (Human) | PR |
| P0CW01 | TSPY10 | Testis-specific Y-encoded protein 10 | Homo sapiens (Human) | PR |
| A6NKD2 | TSPY2 | Testis-specific Y-encoded protein 2 | Homo sapiens (Human) | PR |
| Q8N831 | TSPYL6 | Testis-specific Y-encoded-like protein 6 | Homo sapiens (Human) | PR |
| Q9H2G4 | TSPYL2 | Testis-specific Y-encoded-like protein 2 | Homo sapiens (Human) | PR |
| P0DME0 | SETSIP | Protein SETSIP | Homo sapiens (Human) | SS |
| Q01105 | SET | Protein SET | Homo sapiens (Human) | SS |
| Q9H489 | TSPY26P | Putative testis-specific Y-encoded-like protein 3 | Homo sapiens (Human) | PR |
| Q9R1M3 | Tspy1 | Testis-specific Y-encoded protein 1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRPEGSLTYW | VPERLRQGFC | GVGRAAQALV | CASAKEGTAF | RMEAVQEGAA | GVESEQAALG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EEAVLLLDDI | MAEVEVVAEE | EGLVERREEA | QRAQQAVPGP | GPMTPESALE | ELLAVQVELE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PVNAQARKAF | SRQREKMERR | RKPHLDRRGA | VIQSVPGFWA | NVIANHPQMS | ALITDEDEDM |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LSYMVSLEVE | EEKHPVHLCK | IMLFFRSNPY | FQNKVITKEY | LVNITEYRAS | HSTPIEWYLD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| YEVEAYRRRH | HNSSLNFFNW | FSDHNFAGSN | KIAEILCKDL | WRNPLQYYKR | MKPPEEGTET |
| SGDSQLLS |