P0CV99
Gene name |
TSPY4 |
Protein name |
Testis-specific Y-encoded protein 4 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:728395 |
EC number |
|
Protein Class |
|
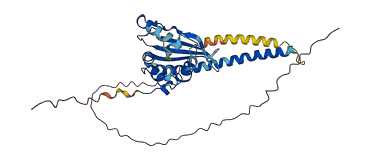
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P0CV99
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P0CV99-F1 | Predicted | AlphaFoldDB |
18 variants for P0CV99
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs879057884 CA337637587 |
10 | R>W | No |
ClinGen Ensembl |
|
|
CA414781682 rs1603139257 |
52 | V>A | No |
ClinGen Ensembl |
|
|
CA414782147 rs1603139262 |
74 | V>E | No |
ClinGen Ensembl |
|
|
rs1603139262 CA414782153 |
74 | V>G | No |
ClinGen Ensembl |
|
|
CA414782268 rs1603139267 |
80 | V>G | No |
ClinGen Ensembl |
|
|
CA414782307 rs1603139270 |
82 | V>G | No |
ClinGen Ensembl |
|
|
CA414782527 rs1603139271 |
94 | E>G | No |
ClinGen Ensembl |
|
|
rs1603139275 CA414782547 |
95 | E>G | No |
ClinGen Ensembl |
|
|
rs1603139282 CA414782876 |
113 | S>A | No |
ClinGen Ensembl |
|
|
CA414783351 rs1556179159 |
137 | S>T | No |
ClinGen Ensembl |
|
|
CA414783409 rs1259762569 |
140 | R>W | No |
ClinGen Ensembl |
|
|
rs1332765786 CA414783639 |
150 | H>Q | No |
ClinGen gnomAD |
|
|
CA414783687 rs1199647885 |
153 | R>C | No |
ClinGen Ensembl |
|
|
rs1257206373 CA414783804 |
159 | Q>P | No |
ClinGen Ensembl |
|
|
CA414785745 rs1556179224 |
243 | W>* | No |
ClinGen Ensembl |
|
|
rs1556179230 CA414785946 |
253 | R>C | No |
ClinGen Ensembl |
|
|
rs1556179264 CA414787350 |
310 | S>F | No |
ClinGen Ensembl |
|
|
CA414787358 rs1603139360 |
311 | Q>* | No |
ClinGen Ensembl |
No associated diseases with P0CV99
No regional properties for P0CV99
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P0CV99 | |||
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| histone binding | Binding to a histone, any of a group of water-soluble proteins found in association with the DNA of eukaryotic or archaeal chromosomes. They are involved in the condensation and coiling of chromosomes during cell division and have also been implicated in gene regulation and DNA replication. They may be chemically modified (methylated, acetlyated and others) to regulate gene transcription. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| gonadal mesoderm development | The process whose specific outcome is the progression of the gonadal mesoderm over time, from its formation to the mature structure. The gonadal mesoderm is the middle layer of the three primary germ layers of the embryo which will go on to form the gonads of the organism. |
| nucleosome assembly | The aggregation, arrangement and bonding together of a nucleosome, the beadlike structural units of eukaryotic chromatin composed of histones and DNA. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O19110 | TSPY1 | Testis-specific Y-encoded protein 1 | Bos taurus (Bovine) | PR |
| Q99457 | NAP1L3 | Nucleosome assembly protein 1-like 3 | Homo sapiens (Human) | PR |
| P55209 | NAP1L1 | Nucleosome assembly protein 1-like 1 | Homo sapiens (Human) | PR |
| Q01534 | TSPY1 | Testis-specific Y-encoded protein 1 | Homo sapiens (Human) | PR |
| P0CV98 | TSPY3 | Testis-specific Y-encoded protein 3 | Homo sapiens (Human) | PR |
| P0CW00 | TSPY8 | Testis-specific Y-encoded protein 8 | Homo sapiens (Human) | PR |
| P0CW01 | TSPY10 | Testis-specific Y-encoded protein 10 | Homo sapiens (Human) | PR |
| A6NKD2 | TSPY2 | Testis-specific Y-encoded protein 2 | Homo sapiens (Human) | PR |
| Q8N831 | TSPYL6 | Testis-specific Y-encoded-like protein 6 | Homo sapiens (Human) | PR |
| Q9H2G4 | TSPYL2 | Testis-specific Y-encoded-like protein 2 | Homo sapiens (Human) | PR |
| P0DME0 | SETSIP | Protein SETSIP | Homo sapiens (Human) | SS |
| Q01105 | SET | Protein SET | Homo sapiens (Human) | SS |
| Q9H489 | TSPY26P | Putative testis-specific Y-encoded-like protein 3 | Homo sapiens (Human) | PR |
| Q9EQU5 | Set | Protein SET | Mus musculus (Mouse) | EV |
| Q9R1M3 | Tspy1 | Testis-specific Y-encoded protein 1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRPEGSLTYR | VPERLRQGFC | GVGRAAQALV | CASAKEGTAF | RMEAVQEGAA | GVESEQAALG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EEAVLLLDDI | MAEVEVVAEV | EVVAEEEGLV | ERREEAQRAQ | QAVPGPGPMT | PESALEELLA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VQVELEPVNA | QARKAFSRQR | EKMERRRKPH | LDRRGAVIQS | VPGFWANVIA | NHPQMSALIT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DEDEDMLSYM | VSLEVEEEKH | PVHLCKIMLF | FRSNPYFQNK | VITKEYLVNI | TEYRASHSTP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IEWYPDYEVE | AYRRRHHNSS | LNFFNWFSDH | NFAGSNKIAE | ILCKDLWRNP | LQYYKRMKPP |
| 310 | |||||
| EEGTETSGDS | QLLS |