P0CE12
Gene name |
sspH2 (STM2241) |
Protein name |
E3 ubiquitin-protein ligase SspH2 |
Names |
RING-type E3 ubiquitin transferase SspH2, Salmonella secreted protein H2, Secreted effector protein sspH2 |
Species |
Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) |
KEGG Pathway |
stm:STM2241 |
EC number |
2.3.2.27: Aminoacyltransferases |
Protein Class |
FAMILY NOT NAMED (PTHR47114) |
Descriptions
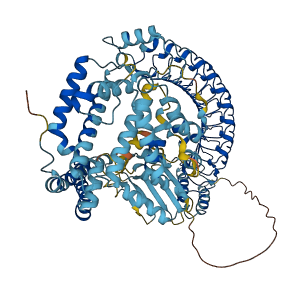
SspH2, consisting of an LRR domain and a Novel E3 ligase (NEL) domain, is an E3 ubiquitin ligase with unique folding. The NEL domain has a catalytic activity that is inhibited by the LRR domain by sequestering the catalytic cysteine residue contained in the NEL domain. The N-terminal domain targets SspH2 to the apical plasma membrane of polarized epithelial cells, which is expected to play a role in releasing the ligase domain for site-specific function.
Autoinhibitory domains (AIDs)
Target domain |
502-712 (Novel E3 ligase, NEL domain) |
Relief mechanism |
Partner binding |
Assay |
Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
References
- Quezada CM et al. (2009) "A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases", Proceedings of the National Academy of Sciences of the United States of America, 106, 4864-9
- Chou YC et al. (2012) "Conserved structural mechanisms for autoinhibition in IpaH ubiquitin ligases", The Journal of biological chemistry, 287, 268-275
Autoinhibited structure
Activated structure

2 structures for P0CE12
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3G06 | X-ray | 190 A | A | 166-783 | PDB |
| AF-P0CE12-F1 | Predicted | AlphaFoldDB |
No variants for P0CE12
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P0CE12 | |||||
No associated diseases with P0CE12
10 regional properties for P0CE12
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Leucine-rich repeat | 243 - 264 | IPR001611-1 |
| repeat | Leucine-rich repeat | 303 - 324 | IPR001611-2 |
| repeat | Leucine-rich repeat | 343 - 364 | IPR001611-3 |
| repeat | Leucine-rich repeat | 383 - 404 | IPR001611-4 |
| repeat | Leucine-rich repeat, typical subtype | 242 - 267 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 302 - 327 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 340 - 367 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 380 - 403 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 420 - 444 | IPR003591-5 |
| domain | Novel E3 ligase domain | 502 - 714 | IPR029487 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.27 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47114 | FAMILY NOT NAMED |
| PANTHER Subfamily | PTHR47114:SF2 | OLIGODENDROCYTE-MYELIN GLYCOPROTEIN |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| host cell cytoplasm | The cytoplasm of a host cell. |
| host cell plasma membrane | The plasma membrane surrounding a host cell. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| catalytic activity | Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| positive regulation of interleukin-8 production | Any process that activates or increases the frequency, rate, or extent of interleukin-8 production. |
| positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | Any process that activates or increases the frequency, rate, or extent of the nucleotide-binding oligomerization domain containing 1 (NOD1) pathway. |
| positive regulation of plant-type hypersensitive response | Any process that activates or increases the frequency, rate or extent of the hypersensitive response in a plant. |
| protein secretion by the type III secretion system | The process in which proteins are transferred into the extracellular milieu or directly into host cells by the bacterial type III secretion system; secretion occurs in a continuous process without the distinct presence of periplasmic intermediates and does not involve proteolytic processing of secreted proteins. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPFHIGSGCL | PATISNRRIY | RIAWSDTPPE | MSSWEKMKEF | FCSTHQTEAL | ECIWTICHPP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AGTTREDVIN | RFELLRTLAY | AGWEESIHSG | QHGENYFCIL | DEDSQEILSV | TLDDAGNYTV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NCQGYSETHR | LTLDTAQGEE | GTGHAEGASG | TFRTSFLPAT | TAPQTPAEYD | AVWSAWRRAA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PAEESRGRAA | VVQKMRACLN | NGNAVLNVGE | SGLTTLPDCL | PAHITTLVIP | DNNLTSLPAL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PPELRTLEVS | GNQLTSLPVL | PPGLLELSIF | SNPLTHLPAL | PSGLCKLWIF | GNQLTSLPVL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PPGLQELSVS | DNQLASLPAL | PSELCKLWAY | NNQLTSLPML | PSGLQELSVS | DNQLASLPTL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PSELYKLWAY | NNRLTSLPAL | PSGLKELIVS | GNRLTSLPVL | PSELKELMVS | GNRLTSLPML |
| 430 | 440 | 450 | 460 | 470 | 480 |
| PSGLLSLSVY | RNQLTRLPES | LIHLSSETTV | NLEGNPLSER | TLQALREITS | APGYSGPIIR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| FDMAGASAPR | ETRALHLAAA | DWLVPAREGE | PAPADRWHMF | GQEDNADAFS | LFLDRLSETE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| NFIKDAGFKA | QISSWLAQLA | EDEALRANTF | AMATEATSSC | EDRVTFFLHQ | MKNVQLVHNA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EKGQYDNDLA | ALVATGREMF | RLGKLEQIAR | EKVRTLALVD | EIEVWLAYQN | KLKKSLGLTS |
| 670 | 680 | 690 | 700 | 710 | 720 |
| VTSEMRFFDV | SGVTVTDLQD | AELQVKAAEK | SEFREWILQW | GPLHRVLERK | APERVNALRE |
| 730 | 740 | 750 | 760 | 770 | 780 |
| KQISDYEETY | RMLSDTELRP | SGLVGNTDAE | RTIGARAMES | AKKTFLDGLR | PLVEEMLGSY |
| LNVQWRRN |