Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
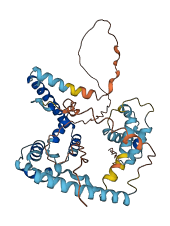
Autoinhibited structure

Activated structure

1 structures for P0CB48
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P0CB48-F1 | Predicted | AlphaFoldDB |
No variants for P0CB48
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P0CB48 | |||||
No associated diseases with P0CB48
5 regional properties for P0CB48
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 513 - 786 | IPR000719 |
| domain | Adenylyl cyclase class-3/4/guanylyl cyclase | 825 - 1037 | IPR001054 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 540 - 783 | IPR001245 |
| domain | Receptor, ligand binding region | 45 - 397 | IPR001828 |
| conserved_site | Adenylyl cyclase class-4/guanylyl cyclase, conserved site | 968 - 991 | IPR018297 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| RNA polymerase I core promoter sequence-specific DNA binding | Binding to a regulatory region composed of the transcription start site and binding sites for transcription factors of the RNA polymerase I transcription machinery. This site is often referred to as the CORE element. In mammalian cells, the CORE element functions in conjunction with the Upstream Control Element (UCE), while in fungi, protozoa, and plants, the CORE element functions without a UCE. |
| RNA polymerase I general transcription initiation factor activity | A general transcription initiation factor activity that contributes to transcription start site selection and transcription initiation of genes transcribed by RNA polymerase I. Factors required for RNA polymerase I transcription initiation include upstream activation factor (UAF), core factor (CF), TATA binding protein (TBP) and RRN3. In all species characterized, RNA polymerase I transcribes a large polycistronic transcript that is processed into several mature rRNAs (3 or 4 depending on the species), including the large subunit rRNA (28S in humans), the small subunit rRNA (18S in humans), as well as one or two additional smaller rRNAs (the 5.8S rRNA in humans). In most species, this large rRNA transcript is the sole product of RNA polymerase I. However there are rare exceptions, such as Trypanosoma brucei, where RNA polymerase I also transcribes certain mRNAs. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| positive regulation of transcription by RNA polymerase I | Any process that activates or increases the frequency, rate or extent of transcription mediated by RNA polymerase I. |
| transcription by RNA polymerase I | The synthesis of RNA from a DNA template by RNA polymerase I (RNAP I), originating at an RNAP I promoter. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPAKDIKVAL | PRSQGHWSNA | DILRLLECME | NNLPYDDNGT | FSSTQSHMDW | GKVAFKNFSG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EMCRLKWLEI | SCSLRKFSTL | KKLVLEAKKC | VKNTNKSQKG | RNHPDFPKRL | LTAYIRFFKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NWPQYSQMYP | GMRSQEVTKI | LSKKYKELPE | QMKQKHIQDF | RKEKQEFEEK | LARFREEHPD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LDQKGKKSDI | CKRVQTKVQK | KVQKNIEEVR | SLPKTDQFFK | KVKFHGEPQK | PPMNGYQKFH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QDSWSSKELQ | HLSLRERMVE | IGRRWQRIPQ | SQKDHYKSQA | ELLQKEYKVE | LDLWLKTLSP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EDYAAYKEST | YAKGKNMAMM | GGPAPSLKQT | DPQSSSAKGL | QEGFGEGQGL | QAAGTEASQT |
| 370 | 380 | 390 | |||
| IWVNCQVSME | PEDNRKKDGE | EEESSNSLDC | SSGEDMEVDV |