P0C1Q2
Gene name |
Pde11a |
Protein name |
Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A |
Names |
EC 3.1.4.35 , EC 3.1.4.53 , cAMP and cGMP phosphodiesterase 11A |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:241489 |
EC number |
3.1.4.35: Phosphoric diester hydrolases |
Protein Class |
|
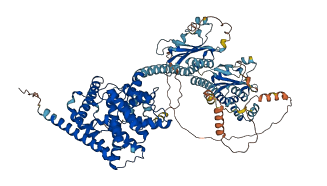
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
295-377 (GAF a domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Zoraghi R et al. (2005) "Structural and functional features in human PDE5A1 regulatory domain that provide for allosteric cGMP binding, dimerization, and regulation", The Journal of biological chemistry, 280, 12051-63
- Francis SH et al. (2011) "Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions", Physiological reviews, 91, 651-90
Autoinhibited structure

Activated structure

1 structures for P0C1Q2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P0C1Q2-F1 | Predicted | AlphaFoldDB |
34 variants for P0C1Q2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388563323 | 59 | S>N | No | EVA | |
| rs3388554254 | 94 | A>V | No | EVA | |
| rs3388554290 | 99 | S>N | No | EVA | |
| rs3388554290 | 99 | S>T | No | EVA | |
| rs3388561248 | 104 | S>C | No | EVA | |
| rs3388556200 | 105 | G>D | No | EVA | |
| rs3388562060 | 159 | R>G | No | EVA | |
| rs3388560895 | 196 | L>I | No | EVA | |
| rs3388559714 | 204 | F>L | No | EVA | |
| rs3391773001 | 209 | V>* | No | EVA | |
| rs3388557810 | 227 | I>N | No | EVA | |
| rs3391832497 | 230 | C>S | No | EVA | |
| rs3388563257 | 271 | S>G | No | EVA | |
| rs3391798518 | 355 | D>V | No | EVA | |
| rs3388563271 | 371 | S>F | No | EVA | |
| rs3388563411 | 397 | F>S | No | EVA | |
| rs3388561220 | 480 | A>T | No | EVA | |
| rs27963339 | 499 | A>T | No | EVA | |
| rs3388563288 | 534 | L>F | No | EVA | |
| rs3388559748 | 546 | L>P | No | EVA | |
| rs3388562047 | 607 | A>T | No | EVA | |
| rs3388561021 | 636 | M>I | No | EVA | |
| rs3388556551 | 678 | A>P | No | EVA | |
| rs3391768786 | 699 | V>E | No | EVA | |
| rs3410289562 | 718 | K>R | No | EVA | |
| rs3391625618 | 780 | Y>N | No | EVA | |
| rs3411762483 | 801 | T>I | No | EVA | |
| rs3388557750 | 852 | T>A | No | EVA | |
| rs3388556545 | 859 | R>G | No | EVA | |
| rs3388557745 | 859 | R>Q | No | EVA | |
| rs3388556189 | 864 | E>V | No | EVA | |
| rs3388552549 | 881 | Y>* | No | EVA | |
| rs3388561080 | 882 | Q>R | No | EVA | |
| rs3388552554 | 909 | H>L | No | EVA |
No associated diseases with P0C1Q2
10 regional properties for P0C1Q2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FERM domain | 35 - 355 | IPR000299 |
| domain | Protein kinase domain | 422 - 680 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 422 - 676 | IPR001245 |
| domain | Focal adhesion kinase, targeting (FAT) domain | 917 - 1048 | IPR005189 |
| active_site | Tyrosine-protein kinase, active site | 542 - 554 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 428 - 454 | IPR017441 |
| domain | FERM central domain | 135 - 250 | IPR019748 |
| domain | Band 4.1 domain | 31 - 258 | IPR019749 |
| domain | Tyrosine-protein kinase, catalytic domain | 422 - 676 | IPR020635 |
| domain | FAK1/PYK2, FERM domain C-lobe | 254 - 364 | IPR041784 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.4.35 | Phosphoric diester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| perikaryon | The portion of the cell soma (neuronal cell body) that excludes the nucleus. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| 3',5'-cyclic-AMP phosphodiesterase activity | Catalysis of the reaction |
| 3',5'-cyclic-GMP phosphodiesterase activity | Catalysis of the reaction |
| cGMP binding | Binding to cGMP, the nucleotide cyclic GMP (guanosine 3',5'-cyclophosphate). |
| cGMP-stimulated cyclic-nucleotide phosphodiesterase activity | Catalysis of the reaction |
| cyclic-nucleotide phosphodiesterase activity | Catalysis of the reaction |
| metal ion binding | Binding to a metal ion. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cAMP-mediated signaling | Any intracellular signal transduction in which the signal is passed on within the cell via cyclic AMP (cAMP). Includes production of cAMP, and downstream effectors that further transmit the signal within the cell. |
| metabolic process | The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation. |
| negative regulation of cAMP-mediated signaling | Any process which stops, prevents, or reduces the frequency, rate or extent of cAMP-mediated signaling. |
| negative regulation of cGMP-mediated signaling | Any process that decreases the rate, frequency or extent of cGMP-mediated signaling. |
32 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q28156 | PDE5A | cGMP-specific 3',5'-cyclic phosphodiesterase | Bos taurus (Bovine) | SS |
| P23439 | PDE6B | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Bos taurus (Bovine) | PR |
| P52731 | PDE6C | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' | Gallus gallus (Chicken) | PR |
| H2QL32 | PDE9A | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Pan troglodytes (Chimpanzee) | PR |
| Q9W4T4 | dnc | 3',5'-cyclic-AMP phosphodiesterase, isoform I | Drosophila melanogaster (Fruit fly) | SS |
| Q9VJ79 | Pde11 | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11 | Drosophila melanogaster (Fruit fly) | SS |
| O76083 | PDE9A | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Homo sapiens (Human) | PR |
| Q07343 | PDE4B | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | Homo sapiens (Human) | EV SS |
| Q08499 | PDE4D | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | Homo sapiens (Human) | EV |
| O76074 | PDE5A | cGMP-specific 3',5'-cyclic phosphodiesterase | Homo sapiens (Human) | EV |
| Q08493 | PDE4C | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Homo sapiens (Human) | EV SS |
| P51160 | PDE6C | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' | Homo sapiens (Human) | PR |
| P27815 | PDE4A | cAMP-specific 3',5'-cyclic phosphodiesterase 4A | Homo sapiens (Human) | EV SS |
| P35913 | PDE6B | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Homo sapiens (Human) | PR |
| Q9Y233 | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Homo sapiens (Human) | PR |
| Q9HCR9 | PDE11A | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Homo sapiens (Human) | SS |
| Q8CG03 | Pde5a | cGMP-specific 3',5'-cyclic phosphodiesterase | Mus musculus (Mouse) | SS |
| O89084 | Pde4a | 3',5'-cyclic-AMP phosphodiesterase 4A | Mus musculus (Mouse) | SS |
| Q3UEI1 | Pde4c | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Mus musculus (Mouse) | PR |
| Q01063 | Pde4d | 3',5'-cyclic-AMP phosphodiesterase 4D | Mus musculus (Mouse) | SS |
| O70628 | Pde9a | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Mus musculus (Mouse) | PR |
| Q8CA95 | Pde10a | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Mus musculus (Mouse) | PR |
| P23440 | Pde6b | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta | Mus musculus (Mouse) | PR |
| Q8QZV1 | Pde9a | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A | Rattus norvegicus (Rat) | PR |
| P14646 | Pde4b | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | Rattus norvegicus (Rat) | SS |
| O54735 | Pde5a | cGMP-specific 3',5'-cyclic phosphodiesterase | Rattus norvegicus (Rat) | SS |
| P14644 | Pde4c | cAMP-specific 3',5'-cyclic phosphodiesterase 4C | Rattus norvegicus (Rat) | PR |
| P54748 | Pde4a | 3',5'-cyclic-AMP phosphodiesterase 4A | Rattus norvegicus (Rat) | SS |
| Q9QYJ6 | Pde10a | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | Rattus norvegicus (Rat) | PR |
| P14270 | Pde4d | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | Rattus norvegicus (Rat) | PR |
| Q8VID6 | Pde11a | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A | Rattus norvegicus (Rat) | SS |
| Q22000 | pde-4 | Probable 3',5'-cyclic phosphodiesterase pde-4 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAASRLDFGE | VETFLDRHPE | LFEDYLMRKG | KQELVDKWLQ | RHTSGQGASS | LRPALAGASS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LAQSNAKGSP | GIGGGAGPQG | SAHSHPTPGG | GESAGVPLSP | SWASGSRGDG | SLQRRASQKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LRKSFARSKA | IHVNRTYDEQ | VTSRAQEPLS | SVRRRALLRK | ASSLPPTTAH | ILSALLESRV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NLPQYPPTAI | DYKCHLKKHN | ERQFFLELVK | DISNDLDLTS | LSYKILIFVC | LMVDADRCSL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FLVEGAAAGK | KTLVSKFFDV | HAGTPLLPCS | STENSNEVQV | PWGKGIIGYV | GEHGETVNIP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DAYQDRRFND | EIDKLTGYKT | KSLLCMPIRN | SDGEIIGVAQ | AINKVPEGAP | FTEDDEKVMQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| MYLPFCGIAI | SNAQLFAASR | KEYERSRALL | EVVNDLFEEQ | TDLEKIVKKI | MHRAQTLLKC |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ERCSVLLLED | IESPVVKFTK | SFELMSPKCS | ADAENSFKES | VEKSSYSDWL | INNSIAELVA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| STGLPVNVSD | AYQDPRFDAE | ADQISGFHIR | SVLCVPIWNS | NHQIIGVAQV | LNRLDGKPFD |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DADQRLFEAF | VIFCGLGINN | TIMYDQVKKS | WAKQSVALDV | LSYHATCSKA | EVDKFKAANI |
| 610 | 620 | 630 | 640 | 650 | 660 |
| PLVSELAIDD | IHFDDFSLDV | DAMITAALRM | FMELGMVQKF | KIDYETLCRW | LLTVRKNYRM |
| 670 | 680 | 690 | 700 | 710 | 720 |
| VLYHNWRHAF | NVCQLMFAML | TTAGFQEILT | EVEILAVIVG | CLCHDLDHRG | TNNAFQAKSD |
| 730 | 740 | 750 | 760 | 770 | 780 |
| SALAQLYGTS | ATLEHHHFNH | AVMILQSEGH | NIFANLSSKE | YSDLMQLLKQ | SILATDLTLY |
| 790 | 800 | 810 | 820 | 830 | 840 |
| FERRTEFFEL | VRKGDYDWSI | TSHRDVFRSM | LMTACDLGAV | TKPWEISRQV | AELVTSEFFE |
| 850 | 860 | 870 | 880 | 890 | 900 |
| QGDRERSELK | LTPSAIFDRN | RKDELPRLQL | EWIDSICMPL | YQALVKVNAK | LKPMLDSVAA |
| 910 | 920 | 930 | |||
| NRRKWEELHQ | KRLQVSAASP | DPASPMVAGE | DRL |