P09661
Gene name |
SNRPA1 |
Protein name |
U2 small nuclear ribonucleoprotein A' |
Names |
U2 snRNP A' |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6627 |
EC number |
|
Protein Class |
|
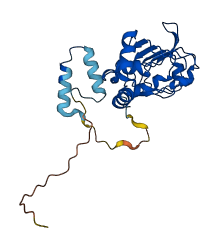
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

31 structures for P09661
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1A9N | X-ray | 238 A | A/C | 1-176 | PDB |
| 5MQF | EM | 590 A | W | 1-255 | PDB |
| 5O9Z | EM | 450 A | z | 1-255 | PDB |
| 5XJC | EM | 360 A | o | 1-255 | PDB |
| 5YZG | EM | 410 A | o | 1-255 | PDB |
| 5Z56 | EM | 510 A | o | 1-255 | PDB |
| 5Z57 | EM | 650 A | o | 1-255 | PDB |
| 5Z58 | EM | 490 A | o | 1-255 | PDB |
| 6AH0 | EM | 570 A | o | 1-255 | PDB |
| 6AHD | EM | 380 A | o | 1-255 | PDB |
| 6FF7 | EM | 450 A | W | 1-255 | PDB |
| 6ICZ | EM | 300 A | o | 1-255 | PDB |
| 6ID0 | EM | 290 A | o | 1-255 | PDB |
| 6ID1 | EM | 286 A | o | 1-255 | PDB |
| 6QDV | EM | 330 A | W | 2-162 | PDB |
| 6QX9 | EM | 328 A | 2A | 1-255 | PDB |
| 6Y53 | EM | 710 A | a | 1-255 | PDB |
| 6Y5Q | EM | 710 A | a | 1-255 | PDB |
| 7A5P | EM | 500 A | W | 1-255 | PDB |
| 7ABG | EM | 780 A | W | 1-255 | PDB |
| 7ABI | EM | 800 A | W | 1-255 | PDB |
| 7EVO | EM | 250 A | F | 1-255 | PDB |
| 7VPX | EM | 300 A | F | 1-255 | PDB |
| 7W59 | EM | 360 A | o | 1-255 | PDB |
| 7W5A | EM | 360 A | o | 1-255 | PDB |
| 7W5B | EM | 430 A | o | 1-255 | PDB |
| 8C6J | EM | 280 A | W | 1-255 | PDB |
| 8CH6 | EM | 590 A | r | 1-255 | PDB |
| 8HK1 | EM | 270 A | F | 1-255 | PDB |
| 8QO9 | EM | 529 A | 2A | 1-255 | PDB |
| AF-P09661-F1 | Predicted | AlphaFoldDB |
144 variants for P09661
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 1 | M>? | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7763347 rs777988186 |
4 | L>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1404929833 CA393967197 |
5 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs772555366 CA7763346 |
5 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393967167 rs1390475867 |
10 | E>K | No |
ClinGen TOPMed |
|
|
CA276343375 rs561721413 CA7763344 |
11 | Q>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1040869856 CA276343370 |
13 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA7763343 rs755005074 |
14 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1243664450 CA393967133 |
15 | Y>H | No |
ClinGen TOPMed |
|
|
rs1447146268 CA393967126 |
16 | T>A | No |
ClinGen gnomAD |
|
|
rs1314344421 CA393967118 |
17 | N>S | No |
ClinGen TOPMed |
|
|
CA7763342 rs753952520 |
20 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA7763341 rs781038951 |
21 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA7763340 rs757152353 |
22 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393967072 rs1400924250 |
25 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA7763338 rs763788242 |
26 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs754665870 CA7763314 |
28 | G>E | No |
ClinGen ExAC |
|
|
rs905473905 CA276343248 |
28 | G>R | No |
ClinGen TOPMed |
|
|
rs1226462085 CA393967034 |
29 | Y>C | No |
ClinGen gnomAD |
|
|
rs1272146544 CA393967037 |
29 | Y>H | No |
ClinGen gnomAD |
|
| TCGA novel | 30 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs138801817 CA7763311 |
32 | P>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs755270186 CA276340915 |
33 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA393966990 rs1443545247 |
36 | N>H | No |
ClinGen gnomAD |
|
|
rs749503765 CA7763305 |
40 | T>K | No |
ClinGen ExAC gnomAD |
|
|
rs749503765 CA7763306 |
40 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs143051052 CA276340882 |
47 | I>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA7763301 rs557125509 |
51 | D>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1421616242 CA393966888 |
51 | D>N | No |
ClinGen gnomAD |
|
|
CA393966883 rs1404755444 |
51 | D>V | No |
ClinGen TOPMed gnomAD |
|
|
rs138420806 CA7763300 |
53 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs947423040 CA276340853 |
58 | D>N | No |
ClinGen TOPMed |
|
|
rs1490445110 CA393966830 |
59 | G>D | No |
ClinGen gnomAD |
|
|
CA7763299 rs748156963 |
59 | G>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 60 | F>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 61 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA276340847 rs1053301275 |
63 | L>S | No |
ClinGen TOPMed gnomAD |
|
|
CA7763298 rs778848364 |
64 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs753518771 CA7763296 |
66 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs1438341100 CA393966778 |
67 | K>R | No |
ClinGen TOPMed |
|
|
CA393966751 rs1157834138 |
71 | V>A | No |
ClinGen gnomAD |
|
|
CA393966741 rs1286980212 |
73 | N>H | No |
ClinGen gnomAD |
|
|
CA393966738 rs1409644499 |
73 | N>S | No |
ClinGen gnomAD |
|
| TCGA novel | 75 | R>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7763295 rs766082491 |
75 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs755543547 CA7763294 |
76 | I>L | No |
ClinGen ExAC gnomAD |
|
|
CA7763284 rs745867854 |
78 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs776784734 CA7763283 |
78 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs745867854 CA393980516 |
78 | R>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 81 | E>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 82 | G>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA276391133 rs768641217 |
83 | L>V | No |
ClinGen Ensembl |
|
|
rs1271985821 CA393980451 |
88 | P>S | No |
ClinGen TOPMed |
|
|
rs1207067390 CA393980439 |
90 | L>V | No |
ClinGen TOPMed |
|
|
rs749129281 CA7763278 |
94 | I>F | No |
ClinGen ExAC gnomAD |
|
|
rs1243754804 CA393980392 |
97 | N>S | No |
ClinGen gnomAD |
|
|
rs142731245 CA7763275 |
99 | S>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs114919170 CA7763273 |
100 | L>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs114919170 CA7763274 |
100 | L>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs369821067 CA393980366 |
101 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7763271 rs369821067 |
101 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA7763242 rs769605360 |
108 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA7763243 rs775327517 |
108 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7763241 rs745669436 |
112 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7763240 rs577233883 |
114 | S>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1244018389 CA393979883 |
118 | L>V | No |
ClinGen gnomAD |
|
| TCGA novel | 119 | S>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA393979769 rs1305290777 |
122 | R>G | No |
ClinGen gnomAD |
|
|
rs772901598 CA7763219 |
124 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA393979702 rs1276985340 |
125 | V>A | No |
ClinGen gnomAD |
|
|
CA7763217 rs747643637 |
125 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA7763216 rs779411044 |
126 | T>I | No |
ClinGen ExAC |
|
|
rs755457351 CA7763215 |
127 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749866603 CA7763214 |
129 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
rs780565103 CA7763213 |
130 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
CA393979591 rs1479198702 |
130 | H>R | No |
ClinGen gnomAD |
|
|
rs1400280744 CA393979582 |
131 | Y>D | No |
ClinGen TOPMed gnomAD |
|
|
CA393979511 rs1464999709 |
133 | L>W | No |
ClinGen gnomAD |
|
|
rs1255731640 CA393979466 |
135 | V>A | No |
ClinGen TOPMed |
|
|
rs756583632 CA7763212 |
136 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA393979462 rs1249280480 |
136 | I>V | No |
ClinGen gnomAD |
|
|
CA393979420 rs1446316109 |
138 | K>E | No |
ClinGen gnomAD |
|
|
rs757317130 CA7763209 |
140 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs767869756 CA7763210 |
140 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs767869756 CA7763211 |
140 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA393979347 rs1596470717 |
142 | V>I | No |
ClinGen Ensembl |
|
|
rs1233769122 CA393979336 |
143 | R>G | No |
ClinGen gnomAD |
|
| TCGA novel | 148 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA276385869 rs908504284 |
151 | K>Q | No |
ClinGen Ensembl |
|
|
CA7763192 rs150710048 |
155 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs1316539546 CA393978396 |
158 | A>S | No |
ClinGen Ensembl |
|
|
rs1459858540 CA393978376 |
160 | K>N | No |
ClinGen gnomAD |
|
|
CA7763191 rs781405747 |
161 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1476596171 CA393978370 |
161 | M>T | No |
ClinGen TOPMed |
|
|
rs1166981227 CA393978350 |
164 | G>S | No |
ClinGen TOPMed |
|
| TCGA novel | 165 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA393978335 rs1166299700 |
166 | R>W | No |
ClinGen gnomAD |
|
|
CA393978291 rs1427468833 |
173 | D>N | No |
ClinGen gnomAD |
|
|
rs1413812521 CA393978279 |
174 | I>T | No |
ClinGen gnomAD |
|
|
CA276385184 rs967280716 |
175 | A>T | No |
ClinGen TOPMed |
|
|
CA393978262 rs1396580511 |
177 | R>K | No |
ClinGen TOPMed |
|
|
rs150109462 CA276385157 |
180 | T>I | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs186863613 CA7763161 |
182 | N>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1002602341 CA276384705 |
183 | P>L | No |
ClinGen TOPMed |
|
|
rs761721002 CA393978201 |
184 | G>A | No |
ClinGen ExAC gnomAD |
|
|
CA7763160 rs761721002 |
184 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs527750619 CA393978189 |
186 | G>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs527750619 CA7763159 |
186 | G>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs763834626 CA7763158 |
187 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA276384698 rs866869396 |
188 | P>S | No |
ClinGen gnomAD |
|
|
rs531907281 CA7763157 COSM959677 |
189 | T>A | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA7763153 rs142009828 |
195 | G>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA393978131 rs142009828 |
195 | G>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 196 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1426191744 CA393978129 |
196 | P>T | No |
ClinGen gnomAD |
|
|
CA7763150 COSM1708589 rs773240837 |
197 | S>F | skin [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA7763149 rs772297229 |
198 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA7763147 rs147560644 |
200 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 200 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7763148 rs748428177 |
200 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA393978100 rs1474091788 |
201 | V>I | No |
ClinGen TOPMed |
|
|
CA393978082 rs1418348854 |
203 | A>V | No |
ClinGen TOPMed |
|
|
CA7763123 rs780921357 |
208 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA276383963 rs755978201 |
209 | A>T | No |
ClinGen Ensembl |
|
|
rs1231250957 CA393978016 |
211 | A>V | No |
ClinGen TOPMed |
|
|
rs1396477329 CA393977998 |
215 | A>T | No |
ClinGen gnomAD |
|
|
CA393977995 rs1381110043 |
215 | A>V | No |
ClinGen gnomAD |
|
|
CA393977977 rs1375110306 |
218 | E>K | No |
ClinGen gnomAD |
|
|
CA276383958 rs943781712 |
219 | R>K | No |
ClinGen Ensembl |
|
|
rs1463642202 CA393977951 |
222 | G>R | No |
ClinGen gnomAD |
|
| TCGA novel | 224 | L>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7763118 rs752376277 |
226 | S>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1567124067 CA393977890 |
231 | G>D | No |
ClinGen Ensembl |
|
|
CA7763117 rs373922948 |
233 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1258736102 CA393977873 |
233 | E>K | No |
ClinGen TOPMed |
|
|
CA7763116 rs145302917 |
234 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA gnomAD |
|
rs753401813 CA7763115 |
234 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1567122671 CA393977173 |
243 | E>K | No |
ClinGen Ensembl |
|
|
CA7763094 rs141391407 |
244 | E>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs991548658 CA276380484 |
247 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs762095184 CA7763093 |
249 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393977081 rs1413188935 |
250 | T>K | No |
ClinGen gnomAD |
|
|
CA7763092 rs751930111 |
252 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA393977043 rs1473049355 |
254 | G>R | No |
ClinGen Ensembl |
No associated diseases with P09661
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| catalytic step 2 spliceosome | A spliceosomal complex that contains three snRNPs, including U5, bound to a splicing intermediate in which the first catalytic cleavage of the 5' splice site has occurred. The precise subunit composition differs significantly from that of the catalytic step 1, or activated, spliceosome, and includes many proteins in addition to those found in the associated snRNPs. |
| nuclear body | Extra-nucleolar nuclear domains usually visualized by confocal microscopy and fluorescent antibodies to specific proteins. |
| nuclear speck | A discrete extra-nucleolar subnuclear domain, 20-50 in number, in which splicing factors are seen to be localized by immunofluorescence microscopy. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| small nuclear ribonucleoprotein complex | A ribonucleoprotein complex that contains at least one RNA of the small nuclear RNA (snRNA) class and as well as its associated proteins. These are typically named after the snRNA(s) they contain, e.g. U1 snRNP, U4/U6 snRNP, or 7SK snRNP. Many, of these complexes become part of the spliceosome involved in splicing of nuclear mRNAs. Others are involved in regulation of transcription elongation or 3'-end processing of replication-dependent histone pre-mRNAs. |
| spliceosomal complex | Any of a series of ribonucleoprotein complexes that contain snRNA(s) and small nuclear ribonucleoproteins (snRNPs), and are formed sequentially during the spliceosomal splicing of one or more substrate RNAs, and which also contain the RNA substrate(s) from the initial target RNAs of splicing, the splicing intermediate RNA(s), to the final RNA products. During cis-splicing, the initial target RNA is a single, contiguous RNA transcript, whether mRNA, snoRNA, etc., and the released products are a spliced RNA and an excised intron, generally as a lariat structure. During trans-splicing, there are two initial substrate RNAs, the spliced leader RNA and a pre-mRNA. |
| U2 snRNP | A ribonucleoprotein complex that contains small nuclear RNA U2, a heptameric ring of Sm proteins, as well as several proteins that are unique to the U2 snRNP, most of which remain associated with the U2 snRNA both while the U2 snRNP is free or assembled into a series of spliceosomal complexes. |
| U2-type catalytic step 2 spliceosome | A spliceosomal complex that contains the U2, U5 and U6 snRNPs bound to a splicing intermediate in which the first catalytic cleavage of the 5' splice site has occurred. The precise subunit composition differs significantly from that of the catalytic step 1, or activated, spliceosome, and includes many proteins in addition to those found in the U2, U5 and U6 snRNPs. |
| U2-type precatalytic spliceosome | A spliceosomal complex that is formed by the recruitment of the preassembled U4/U6.U5 tri-snRNP to the prespliceosome. Although all 5 snRNPs are present, the precatalytic spliceosome is catalytically inactive. The precatalytic spliceosome includes many proteins in addition to those found in the U1, U2 and U4/U6.U5 snRNPs. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| U2 snRNA binding | Binding to a U2 small nuclear RNA (U2 snRNA). |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| mRNA splicing, via spliceosome | The joining together of exons from one or more primary transcripts of messenger RNA (mRNA) and the excision of intron sequences, via a spliceosomal mechanism, so that mRNA consisting only of the joined exons is produced. |
| RNA splicing | The process of removing sections of the primary RNA transcript to remove sequences not present in the mature form of the RNA and joining the remaining sections to form the mature form of the RNA. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
| U2-type prespliceosome assembly | The aggregation, arrangement and bonding together of a set of components to form an U2-type prespliceosome. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVKLTAELIE | QAAQYTNAVR | DRELDLRGYK | IPVIENLGAT | LDQFDAIDFS | DNEIRKLDGF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PLLRRLKTLL | VNNNRICRIG | EGLDQALPCL | TELILTNNSL | VELGDLDPLA | SLKSLTYLSI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LRNPVTNKKH | YRLYVIYKVP | QVRVLDFQKV | KLKERQEAEK | MFKGKRGAQL | AKDIARRSKT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FNPGAGLPTD | KKKGGPSPGD | VEAIKNAIAN | ASTLAEVERL | KGLLQSGQIP | GRERRSGPTD |
| 250 | |||||
| DGEEEMEEDT | VTNGS |