P08134
Gene name |
RHOC (ARH9, ARHC) |
Protein name |
Rho-related GTP-binding protein RhoC |
Names |
Rho cDNA clone 9, h9 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:389 |
EC number |
|
Protein Class |
|
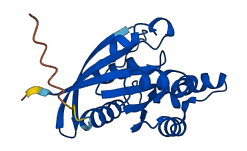
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for P08134
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1Z2C | X-ray | 300 A | A/C | 1-193 | PDB |
| 2GCN | X-ray | 185 A | A | 1-181 | PDB |
| 2GCO | X-ray | 140 A | A/B | 1-181 | PDB |
| 2GCP | X-ray | 215 A | A | 1-181 | PDB |
| AF-P08134-F1 | Predicted | AlphaFoldDB |
122 variants for P08134
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1204217869 CA341669604 |
3 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
CA341669605 rs1204217869 |
3 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA341669601 rs1484661530 |
3 | A>V | No |
ClinGen gnomAD |
|
|
rs549460700 CA1010505 |
4 | I>F | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1010504 rs775884207 |
4 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA28961529 rs549460700 |
4 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs751438534 CA1010503 |
5 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA341669593 rs1341176142 |
5 | R>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 6 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1804292 CA28961498 |
7 | K>N | No |
ClinGen gnomAD |
|
|
rs1570832100 CA341669567 |
9 | V>G | No |
ClinGen Ensembl |
|
|
rs777122070 CA1010501 |
11 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs777122070 CA341669559 |
11 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1279883584 CA341669534 |
15 | A>T | No |
ClinGen gnomAD |
|
|
CA28961475 rs774806474 |
16 | C>F | No |
ClinGen Ensembl |
|
|
CA341669521 rs1368561952 |
17 | G>R | No |
ClinGen gnomAD |
|
|
rs1570832032 CA341669505 |
19 | T>P | No |
ClinGen Ensembl |
|
|
CA1010498 rs532814642 |
20 | C>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1010497 rs778283490 |
22 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs754465217 CA1010496 |
23 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs779724269 CA1010494 |
31 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1306776701 CA341669409 |
33 | V>I | No |
ClinGen gnomAD |
|
| TCGA novel | 35 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341669393 rs1373957183 |
35 | V>D | No |
ClinGen gnomAD |
|
|
rs867290320 CA28961425 |
36 | P>L | No |
ClinGen Ensembl |
|
|
rs751468245 CA1010489 |
38 | V>A | No |
ClinGen ExAC |
|
|
rs1225428332 CA341669331 |
44 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
CA341669318 rs1445187406 |
46 | I>N | No |
ClinGen gnomAD |
|
|
CA341669320 rs1279096579 |
46 | I>V | No |
ClinGen gnomAD |
|
|
CA28961398 rs113742657 |
48 | V>A | No |
ClinGen Ensembl |
|
|
CA1010487 rs775687775 |
49 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1010485 rs142648390 |
50 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1051591442 CA28961379 |
51 | K>R | No |
ClinGen Ensembl |
|
|
CA341669277 rs1325886799 |
52 | Q>R | No |
ClinGen gnomAD |
|
|
rs1570830882 CA341669257 |
53 | V>G | No |
ClinGen Ensembl |
|
|
rs1291714036 CA341669243 |
56 | A>S | No |
ClinGen gnomAD |
|
| TCGA novel | 63 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 65 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs760987953 CA1010458 |
66 | Y>S | No |
ClinGen ExAC gnomAD |
|
|
COSM893969 CA341669155 rs1285399896 |
68 | R>Q | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs772515757 CA1010456 |
70 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA28961044 rs768341873 |
79 | V>I | No |
ClinGen Ensembl |
|
|
rs111933338 CA1010452 |
82 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA341669042 rs1007576553 |
86 | I>L | No |
ClinGen gnomAD |
|
|
rs780741469 CA1010451 |
86 | I>N | No |
ClinGen ExAC gnomAD |
|
|
CA28961035 rs1007576553 |
86 | I>V | No |
ClinGen gnomAD |
|
|
CA341669021 rs1475122235 |
89 | P>S | No |
ClinGen gnomAD |
|
| TCGA novel | 90 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341668996 rs1196046097 |
92 | L>P | No |
ClinGen gnomAD |
|
|
CA28960820 rs982075258 |
95 | I>F | No |
ClinGen TOPMed |
|
|
CA341668966 rs982075258 |
95 | I>V | No |
ClinGen TOPMed |
|
|
rs1012573175 CA28960819 |
96 | P>A | No |
ClinGen Ensembl |
|
|
CA1010425 rs779030492 |
99 | W>R | No |
ClinGen ExAC gnomAD |
|
|
rs754946696 CA1010424 |
100 | T>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 107 | C>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1240537712 CA341668819 |
116 | G>V | No |
ClinGen gnomAD |
|
|
rs750672121 CA1010420 |
119 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs367869187 CA28960787 |
120 | D>E | No |
ClinGen ESP |
|
|
VAR_051974 CA28960788 rs11538959 |
120 | D>H | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs969698119 CA341668789 |
121 | L>M | No |
ClinGen TOPMed gnomAD |
|
|
rs969698119 CA28960786 |
121 | L>V | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 124 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341668741 rs1242466017 |
127 | T>I | No |
ClinGen TOPMed |
|
|
rs1374625555 CA341668695 |
134 | M>T | No |
ClinGen gnomAD |
|
|
CA341668660 rs1354608494 |
137 | E>* | No |
ClinGen TOPMed |
|
|
CA341668658 rs1448612311 |
137 | E>G | No |
ClinGen TOPMed |
|
|
rs1159815720 CA341668653 |
138 | P>A | No |
ClinGen gnomAD |
|
|
rs368489891 CA341668648 |
139 | V>I | No |
ClinGen ESP ExAC TOPMed |
|
|
rs368489891 CA1010399 |
139 | V>L | No |
ClinGen ESP ExAC TOPMed |
|
|
CA341668643 rs202016325 |
140 | R>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs147557411 CA28960317 |
140 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1010396 rs147557411 |
140 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1010397 rs202016325 |
140 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1010394 rs760269743 |
143 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA341668618 rs1192411071 |
144 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs749927529 CA1010393 |
144 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA1010392 COSM347133 rs767339526 |
145 | R>W | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA341668597 rs1355603125 |
146 | D>E | No |
ClinGen gnomAD |
|
|
rs761556555 CA1010390 |
146 | D>H | No |
ClinGen ExAC |
|
|
rs935568461 CA28960274 |
147 | M>I | No |
ClinGen gnomAD |
|
|
rs138756182 CA341668583 |
148 | A>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1263606082 CA341668585 |
148 | A>S | No |
ClinGen gnomAD |
|
|
CA1010389 rs138756182 |
148 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs376797998 CA1010387 |
150 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1010386 rs376797998 |
150 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1044096844 COSM291487 CA28960228 |
150 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1299248790 CA341668564 |
151 | I>T | No |
ClinGen gnomAD |
|
|
rs1389297724 CA341668559 |
152 | S>G | No |
ClinGen gnomAD |
|
|
CA341668546 rs1420071448 |
153 | A>V | No |
ClinGen TOPMed |
|
|
CA1010382 rs770929898 |
157 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA341668516 rs770929898 |
157 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA341668494 rs758818970 |
159 | C>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1450806573 CA341668442 |
165 | E>D | No |
ClinGen gnomAD |
|
| TCGA novel | 168 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1010376 rs755424695 |
168 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs779520374 COSM1560092 CA1010377 |
168 | R>W | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1570827142 CA341668402 |
170 | V>G | No |
ClinGen Ensembl |
|
|
CA341668395 rs1429308276 |
171 | F>S | No |
ClinGen TOPMed |
|
|
rs767143479 CA1010374 |
173 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1216853115 CA341668379 |
173 | M>V | No |
ClinGen gnomAD |
|
| TCGA novel | 174 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341668364 rs1370669974 |
175 | T>A | No |
ClinGen gnomAD |
|
|
rs1304057988 CA341668353 |
176 | R>L | No |
ClinGen gnomAD |
|
|
CA28960130 rs766734264 |
176 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA341668330 rs1400178845 |
179 | L>F | No |
ClinGen gnomAD |
|
|
CA1010369 rs775128679 |
180 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1010367 rs759393969 |
182 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs776490490 CA1010366 |
182 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771017578 CA1010364 |
186 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA1010363 rs11538960 |
186 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1010361 rs766896156 |
187 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779142435 CA341668267 |
187 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779142435 CA1010359 |
187 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA1010360 rs766896156 |
187 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1010358 rs755304944 |
188 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs1200021693 CA341668261 |
188 | R>K | No |
ClinGen gnomAD |
|
|
rs2230329 CA28960090 |
189 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA341668236 rs1570826836 |
191 | P>R | No |
ClinGen Ensembl |
|
|
CA341668229 rs1570826825 |
192 | I>V | No |
ClinGen Ensembl |
|
| TCGA novel | 194 | L>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA341668211 rs1171938191 |
194 | L>R | No |
ClinGen gnomAD |
|
|
rs780659247 CA1010356 |
194 | L>S | No |
ClinGen ExAC gnomAD |
1 associated diseases with P08134
[MIM: 187900]: Bleeding disorder, platelet-type, 17 (BDPLT17)
An autosomal dominant disorder characterized by increased bleeding tendency due to platelet dysfunction, and associated with macrothrombocytopenia and red cell anisopoikilocytosis. Platelets appear abnormal on light microscopy, while electron microscopy shows a heterogeneous decrease of alpha granules within platelets. Bone marrow biopsy shows increased numbers of abnormal megakaryocytes, suggesting a defect in megakaryopoiesis and platelet production. The severity of bleeding is variable with some affected individuals experiencing spontaneous bleeding while other exhibit only abnormal bleeding with surgery. {ECO:0000269|PubMed:23927492, ECO:0000269|PubMed:24325358}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal dominant disorder characterized by increased bleeding tendency due to platelet dysfunction, and associated with macrothrombocytopenia and red cell anisopoikilocytosis. Platelets appear abnormal on light microscopy, while electron microscopy shows a heterogeneous decrease of alpha granules within platelets. Bone marrow biopsy shows increased numbers of abnormal megakaryocytes, suggesting a defect in megakaryopoiesis and platelet production. The severity of bleeding is variable with some affected individuals experiencing spontaneous bleeding while other exhibit only abnormal bleeding with surgery. {ECO:0000269|PubMed:23927492, ECO:0000269|PubMed:24325358}. Note=The disease is caused by variants affecting the gene represented in this entry.
6 regional properties for P08134
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Zinc finger C2H2-type | 163 - 191 | IPR013087-1 |
| domain | Zinc finger C2H2-type | 192 - 219 | IPR013087-2 |
| domain | Zinc finger C2H2-type | 220 - 247 | IPR013087-3 |
| domain | Zinc finger C2H2-type | 248 - 275 | IPR013087-4 |
| domain | Zinc finger C2H2-type | 276 - 303 | IPR013087-5 |
| domain | Zinc finger C2H2-type | 304 - 330 | IPR013087-6 |
Functions
12 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cleavage furrow | The cleavage furrow is a plasma membrane invagination at the cell division site. The cleavage furrow begins as a shallow groove and eventually deepens to divide the cytoplasm. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| stereocilium | An actin-based protrusion from the apical surface of auditory and vestibular hair cells and of neuromast cells. These protrusions are supported by a bundle of cross-linked actin filaments (an actin cable), oriented such that the plus (barbed) ends are at the tip of the protrusion, capped by a tip complex which bridges to the plasma. Bundles of stereocilia act as mechanosensory organelles. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| apical junction assembly | The formation of an apical junction, a functional unit located near the cell apex at the points of contact between epithelial cells composed of the tight junction, the zonula adherens junction and the desmosomes, by the aggregation, arrangement and bonding together of its constituents. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cortical cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures in the cell cortex, i.e. just beneath the plasma membrane. |
| establishment or maintenance of cell polarity | Any cellular process that results in the specification, formation or maintenance of anisotropic intracellular organization or cell growth patterns. |
| mitotic cytokinesis | A cell cycle process that results in the division of the cytoplasm of a cell after mitosis, resulting in the separation of the original cell into two daughter cells. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of I-kappaB kinase/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of I-kappaB kinase/NF-kappaB signaling. |
| positive regulation of lipase activity | Any process that increases the frequency, rate or extent of lipase activity, the hydrolysis of a lipid or phospholipid. |
| positive regulation of protein-containing complex assembly | Any process that activates or increases the frequency, rate or extent of protein complex assembly. |
| positive regulation of stress fiber assembly | Any process that activates or increases the frequency, rate or extent of the assembly of a stress fiber, a bundle of microfilaments and other proteins found in fibroblasts. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| skeletal muscle satellite cell migration | The orderly movement of a skeletal muscle satellite cell from one site to another. Migration of these cells is a key step in the process of growth and repair of skeletal muscle cells. |
| small GTPase mediated signal transduction | The series of molecular signals in which a small monomeric GTPase relays a signal. |
| wound healing, spreading of cells | The migration of a cell along or through a wound gap that contributes to the reestablishment of a continuous surface. |
36 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q17QI8 | Rhov | Rho-related GTP-binding protein RhoV | Bos taurus (Bovine) | PR |
| Q2HJ68 | RND1 | Rho-related GTP-binding protein Rho6 | Bos taurus (Bovine) | PR |
| P61585 | RHOA | Transforming protein RhoA | Bos taurus (Bovine) | PR |
| Q3ZBW5 | RHOB | Rho-related GTP-binding protein RhoB | Bos taurus (Bovine) | PR |
| Q1RMJ6 | RHOC | Rho-related GTP-binding protein RhoC | Bos taurus (Bovine) | PR |
| Q9PSX7 | RHOC | Rho-related GTP-binding protein RhoC | Gallus gallus (Chicken) | PR |
| P48148 | Rho1 | Ras-like GTP-binding protein Rho1 | Drosophila melanogaster (Fruit fly) | PR |
| O94955 | RHOBTB3 | Rho-related BTB domain-containing protein 3 | Homo sapiens (Human) | EV |
| O94844 | RHOBTB1 | Rho-related BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| P61586 | RHOA | Transforming protein RhoA | Homo sapiens (Human) | PR |
| P61587 | RND3 | Rho-related GTP-binding protein RhoE | Homo sapiens (Human) | PR |
| P62745 | RHOB | Rho-related GTP-binding protein RhoB | Homo sapiens (Human) | PR |
| Q92730 | RND1 | Rho-related GTP-binding protein Rho6 | Homo sapiens (Human) | PR |
| Q96L33 | RHOV | Rho-related GTP-binding protein RhoV | Homo sapiens (Human) | PR |
| Q8BLR7 | Rnd1 | Rho-related GTP-binding protein Rho6 | Mus musculus (Mouse) | PR |
| P61588 | Rnd3 | Rho-related GTP-binding protein RhoE | Mus musculus (Mouse) | PR |
| Q9DAK3 | Rhobtb1 | Rho-related BTB domain-containing protein 1 | Mus musculus (Mouse) | PR |
| Q9CTN4 | Rhobtb3 | Rho-related BTB domain-containing protein 3 | Mus musculus (Mouse) | SS |
| Q8VDU1 | Rhov | Rho-related GTP-binding protein RhoV | Mus musculus (Mouse) | PR |
| P62746 | Rhob | Rho-related GTP-binding protein RhoB | Mus musculus (Mouse) | PR |
| Q9QUI0 | Rhoa | Transforming protein RhoA | Mus musculus (Mouse) | PR |
| Q62159 | Rhoc | Rho-related GTP-binding protein RhoC | Mus musculus (Mouse) | PR |
| O77683 | RND3 | Rho-related GTP-binding protein RhoE | Sus scrofa (Pig) | PR |
| Q6SA80 | Rnd3 | Rho-related GTP-binding protein RhoE | Rattus norvegicus (Rat) | PR |
| Q9Z1Y0 | Rhov | Rho-related GTP-binding protein RhoV | Rattus norvegicus (Rat) | PR |
| P61589 | Rhoa | Transforming protein RhoA | Rattus norvegicus (Rat) | PR |
| P62747 | Rhob | Rho-related GTP-binding protein RhoB | Rattus norvegicus (Rat) | PR |
| Q9SSX0 | RAC1 | Rac-like GTP-binding protein 1 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z808 | RAC3 | Rac-like GTP-binding protein 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q68Y52 | RAC2 | Rac-like GTP-binding protein 2 | Oryza sativa subsp japonica (Rice) | PR |
| Q67VP4 | RAC4 | Rac-like GTP-binding protein 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q22038 | rho-1 | Ras-like GTP-binding protein rhoA | Caenorhabditis elegans | PR |
| Q9SU67 | ARAC8 | Rac-like GTP-binding protein ARAC8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82480 | ARAC7 | Rac-like GTP-binding protein ARAC7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38903 | ARAC2 | Rac-like GTP-binding protein ARAC2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82481 | ARAC10 | Rac-like GTP-binding protein ARAC10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAIRKKLVI | VGDGACGKTC | LLIVFSKDQF | PEVYVPTVFE | NYIADIEVDG | KQVELALWDT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AGQEDYDRLR | PLSYPDTDVI | LMCFSIDSPD | SLENIPEKWT | PEVKHFCPNV | PIILVGNKKD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LRQDEHTRRE | LAKMKQEPVR | SEEGRDMANR | ISAFGYLECS | AKTKEGVREV | FEMATRAGLQ |
| 190 | |||||
| VRKNKRRRGC | PIL |