P07199
Gene name |
CENPB |
Protein name |
Major centromere autoantigen B |
Names |
Centromere protein B, CENP-B |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1059 |
EC number |
|
Protein Class |
|
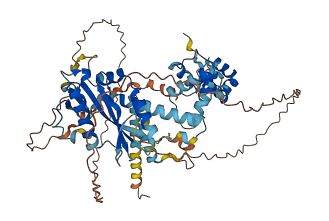
Descriptions
CENP-B Interacts with centromeric heterochromatin in chromosomes and binds to a specific 17 bp subset of alphoid satellite DNA, called the CENP-B box. This protein possesses intrinsically disordered regions (IDRs) with large negative charge, some of which involve a consecutive sequence of aspartate (D) or glutamate (E) residues, known as D/E repeats. These D/E repeats can cause autoinhibition through intramolecular electrostatic interaction with HMG boxes and modulate binding to DNA. This autoinhibited state can transition into the uninhibited complex with DNA through an electrostatically driven induced-fit process, which accelerates the target DNA search kinetics in the presence of non-functional high-affinity ligands ('decoys').
Autoinhibitory domains (AIDs)
Target domain |
1-136 (DNA-binding domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

5 structures for P07199
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1BW6 | NMR | - | A | 1-56 | PDB |
| 1HLV | X-ray | 250 A | A | 1-129 | PDB |
| 1UFI | X-ray | 165 A | A/B/C/D | 540-599 | PDB |
| 6KDR | X-ray | 211 A | D/E | 2-10 | PDB |
| AF-P07199-F1 | Predicted | AlphaFoldDB |
438 variants for P07199
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs768406404 | 3 | P>R | No |
ExAC gnomAD |
|
| rs267605931 | 3 | P>S | No | Ensembl | |
| rs1237739956 | 11 | R>Q | No | gnomAD | |
| rs1265462304 | 11 | R>W | No | gnomAD | |
| rs1345240536 | 21 | E>D | No | gnomAD | |
| rs1407244321 | 22 | E>D | No | gnomAD | |
| rs780512777 | 22 | E>K | No |
ExAC gnomAD |
|
| rs905224989 | 25 | D>N | No |
TOPMed gnomAD |
|
| rs1347102548 | 27 | R>L | No | gnomAD | |
| TCGA novel | 28 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763387256 | 34 | R>H | No |
ExAC gnomAD |
|
| rs1434795060 | 36 | N>S | No | gnomAD | |
| COSM4964654 | 46 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770909940 | 47 | K>N | No |
ExAC gnomAD |
|
| TCGA novel | 50 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1386081685 | 50 | R>S | No |
TOPMed gnomAD |
|
| rs774130454 | 51 | A>S | No |
ExAC gnomAD |
|
| rs1381090845 | 52 | I>L | No | gnomAD | |
| rs1321353770 | 54 | A>V | No | gnomAD | |
| COSM3799529 | 56 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs145734304 | 58 | K>N | No |
ESP ExAC gnomAD |
|
| TCGA novel | 59 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1568495766 | 60 | G>R | No | Ensembl | |
| rs769595472 | 61 | V>M | No |
ExAC gnomAD |
|
| rs745496146 | 62 | A>P | No |
ExAC gnomAD |
|
| rs745496146 | 62 | A>S | No |
ExAC gnomAD |
|
| rs756692439 | 63 | S>P | No |
ExAC gnomAD |
|
| rs533106029 | 64 | T>I | No |
1000Genomes ExAC gnomAD |
|
| rs202224991 | 64 | T>P | No |
ExAC gnomAD |
|
| TCGA novel | 69 | N>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753269687 | 70 | K>R | No |
ExAC gnomAD |
|
| rs1169850506 | 76 | K>R | No | TOPMed | |
| COSM723696 | 77 | L>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1380781333 | 78 | E>K | No |
TOPMed gnomAD |
|
| rs1380781333 | 78 | E>Q | No |
TOPMed gnomAD |
|
| rs1338929899 | 79 | G>S | No | gnomAD | |
| COSM1471464 | 81 | L>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1167131221 | 83 | A>T | No | gnomAD | |
| rs1168609954 | 88 | I>M | No | gnomAD | |
| rs1391956615 | 88 | I>T | No | gnomAD | |
| rs1443469427 | 89 | R>L | No | gnomAD | |
| rs867843060 | 94 | P>A | No | gnomAD | |
| rs1250192663 | 94 | P>R | No | gnomAD | |
| rs867843060 | 94 | P>S | No | gnomAD | |
| rs1367525260 | 96 | K>R | No | TOPMed | |
| rs1194468678 | 97 | G>S | No | gnomAD | |
| TCGA novel | 99 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM723697 | 102 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 109 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745717178 | 113 | M>L | No |
ExAC gnomAD |
|
| TCGA novel | 118 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1372244679 | 121 | G>D | No | gnomAD | |
| rs1362864096 | 133 | V>A | No | gnomAD | |
| rs1362864096 | 133 | V>G | No | gnomAD | |
| rs748658170 | 136 | S>G | No |
ExAC gnomAD |
|
| rs755273196 | 137 | G>S | No |
ExAC gnomAD |
|
| rs531687996 | 139 | A>P | No | 1000Genomes | |
| rs766681316 | 139 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs561079262 | 140 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs561079262 | 140 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1333215305 | 141 | A>G | No | TOPMed | |
| rs1369467816 | 141 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1333215305 | 141 | A>V | No | TOPMed | |
| rs761949795 | 142 | R>C | No |
ExAC gnomAD |
|
| rs975691838 | 146 | A>T | No | TOPMed | |
| rs1352066935 | 147 | A>S | No | gnomAD | |
| rs1352066935 | 147 | A>T | No | gnomAD | |
| rs542584402 | 147 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776448311 | 148 | P>R | No |
ExAC gnomAD |
|
| rs1363667913 | 148 | P>S | No |
TOPMed gnomAD |
|
| TCGA novel | 149 | R>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773064269 | 149 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs771677387 | 149 | R>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 149 | R>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773064269 | 149 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs747708976 | 150 | T>P | No |
ExAC gnomAD |
|
| rs1413132890 | 151 | P>S | No | gnomAD | |
| rs1479704969 | 153 | A>E | No | gnomAD | |
| rs1248979638 | 155 | A>S | No | gnomAD | |
| rs1248979638 | 155 | A>T | No | gnomAD | |
| rs769161961 | 157 | P>A | No |
ExAC gnomAD |
|
| rs572112095 | 157 | P>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756496687 | 159 | A>E | No |
ExAC gnomAD |
|
| rs780404896 | 159 | A>T | No |
ExAC gnomAD |
|
| rs1236440118 | 161 | P>L | No | TOPMed | |
| rs1257221408 | 162 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1268479257 | 163 | E>K | No | gnomAD | |
| rs894008394 | 166 | G>D | No |
TOPMed gnomAD |
|
| rs781437027 | 167 | G>E | No |
ExAC gnomAD |
|
| rs750641264 | 167 | G>W | No |
ExAC gnomAD |
|
| rs201262188 | 168 | S>G | No |
ExAC gnomAD |
|
| rs560373821 | 170 | T>I | No | 1000Genomes | |
| rs751590543 | 171 | G>D | No |
ExAC gnomAD |
|
| rs751590543 | 171 | G>V | No |
ExAC gnomAD |
|
| rs1329456527 | 172 | W>C | No |
TOPMed gnomAD |
|
| rs1458443786 | 172 | W>L | No | gnomAD | |
| rs1388925961 | 173 | R>L | No | gnomAD | |
| rs759452919 | 174 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs759452919 | 174 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs753776622 | 175 | R>Q | No |
ExAC gnomAD |
|
| rs200133266 | 177 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1192254824 | 177 | E>Q | No | gnomAD | |
| rs1240384473 | 179 | P>R | No | gnomAD | |
| rs878883694 | 180 | P>L | No | TOPMed | |
| rs1465463217 | 181 | S>P | No | gnomAD | |
| rs771798121 | 187 | A>P | No |
ExAC gnomAD |
|
| COSM4098338 | 187 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1280523528 | 188 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs867023076 | 189 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1294681306 | 191 | V>M | No |
TOPMed gnomAD |
|
| rs140102208 | 193 | S>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs773929504 | 193 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs768060890 | 194 | A>T | No |
ExAC gnomAD |
|
| rs749791585 | 194 | A>V | No |
ExAC gnomAD |
|
| rs780496154 | 198 | S>R | No |
ExAC gnomAD |
|
| TCGA novel | 199 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 201 | Y>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1409852994 | 202 | D>G | No | gnomAD | |
| rs1309939310 | 203 | F>L | No | TOPMed | |
| rs369656252 | 203 | F>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1600367957 | 205 | P>S | No | Ensembl | |
|
TCGA novel rs1600367947 |
206 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs556461613 | 208 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs1484388993 | 209 | A>G | No |
TOPMed gnomAD |
|
| rs1181946654 | 209 | A>S | No |
TOPMed gnomAD |
|
| rs1181946654 | 209 | A>T | No |
TOPMed gnomAD |
|
| rs1484388993 | 209 | A>V | No |
TOPMed gnomAD |
|
| rs777853235 | 213 | G>V | No |
ExAC TOPMed gnomAD |
|
| COSM1307379 | 216 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs897815576 | 218 | P>L | No |
TOPMed gnomAD |
|
| rs897815576 | 218 | P>R | No |
TOPMed gnomAD |
|
| rs1380923882 | 219 | R>C | No |
TOPMed gnomAD |
|
| rs758472789 | 219 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs758472789 | 219 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1380923882 | 219 | R>S | No |
TOPMed gnomAD |
|
| rs1441954716 | 220 | Q>P | No | gnomAD | |
| rs753775625 | 221 | A>D | No |
ExAC gnomAD |
|
| rs1369159816 | 221 | A>P | No |
TOPMed gnomAD |
|
| rs1384665166 | 223 | Q>K | No |
TOPMed gnomAD |
|
| rs766276108 | 224 | R>L | No |
ExAC gnomAD |
|
| COSM4098337 | 225 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760710349 | 226 | S>C | No |
ExAC gnomAD |
|
| rs750199272 | 226 | S>R | No |
ExAC gnomAD |
|
| rs1178809396 | 231 | A>T | No | gnomAD | |
| COSM723698 | 234 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs773929979 | 235 | G>S | No |
ExAC gnomAD |
|
| rs763658114 | 238 | K>R | No |
ExAC gnomAD |
|
| rs1323093092 | 240 | P>A | No | gnomAD | |
| rs1323093092 | 240 | P>T | No | gnomAD | |
| rs1348985910 | 241 | P>L | No | gnomAD | |
| rs1291791291 | 245 | G>S | No | gnomAD | |
| rs1568495268 | 246 | K>R | No | Ensembl | |
| rs770151730 | 247 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs112241514 | 247 | S>P | No | Ensembl | |
| rs771299910 | 251 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs879399143 | 251 | R>H | No | gnomAD | |
| rs1472837172 | 252 | A>E | No |
TOPMed gnomAD |
|
| rs1184111498 | 252 | A>S | No | gnomAD | |
| rs747225829 | 253 | G>R | No |
ExAC gnomAD |
|
| rs747225829 | 253 | G>S | No |
ExAC gnomAD |
|
| rs758481653 | 255 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1026671116 | 256 | G>A | No | Ensembl | |
| TCGA novel | 257 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748126240 | 257 | L>R | No |
ExAC gnomAD |
|
| rs1214608347 | 258 | P>L | No | gnomAD | |
| COSM4098336 | 261 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs867769633 | 263 | A>T | No | gnomAD | |
| rs1299814146 | 265 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1299814146 | 265 | S>Y | No | gnomAD | |
| rs996878878 | 267 | G>S | No | Ensembl | |
| rs1368366384 | 270 | T>I | No | gnomAD | |
| rs1288385037 | 271 | T>I | No |
TOPMed gnomAD |
|
| rs1433115280 | 273 | A>V | No | gnomAD | |
| rs909068515 | 275 | A>G | No | TOPMed | |
| rs372238704 | 276 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs763706617 | 276 | K>R | No |
ExAC gnomAD |
|
| rs774824085 | 279 | K>R | No |
ExAC gnomAD |
|
| rs774824085 | 279 | K>T | No |
ExAC gnomAD |
|
| rs1267382714 | 280 | A>V | No | gnomAD | |
| rs759981081 | 284 | R>* | No |
ExAC gnomAD |
|
| rs759981081 | 284 | R>G | No |
ExAC gnomAD |
|
| rs1209039813 | 286 | A>V | No | gnomAD | |
| rs1323237462 | 287 | A>S | No | gnomAD | |
| rs1448788680 | 287 | A>V | No | TOPMed | |
| rs1254676165 | 288 | E>Q | No | gnomAD | |
| rs781319502 | 290 | R>C | No | gnomAD | |
|
COSM1681513 rs1372115231 |
291 | R>W | ovary [Cosmic] | No |
cosmic curated TOPMed |
| rs865816773 | 296 | A>V | No | Ensembl | |
| rs1367517935 | 297 | G>S | No | gnomAD | |
| rs555520508 | 298 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs868826693 | 298 | R>H | No | gnomAD | |
| rs868826693 | 298 | R>L | No | gnomAD | |
| rs1300889115 | 301 | A>P | No |
TOPMed gnomAD |
|
| rs147156312 | 303 | S>C | No |
ESP ExAC gnomAD |
|
| rs147156312 | 303 | S>Y | No |
ESP ExAC gnomAD |
|
| rs1453532151 | 307 | S>L | No | TOPMed | |
| rs1447125625 | 308 | G>D | No | gnomAD | |
| rs1341362295 | 310 | R>Q | No | gnomAD | |
| rs1188777924 | 310 | R>W | No | gnomAD | |
| rs755165312 | 311 | H>R | No |
ExAC gnomAD |
|
| rs1483829349 | 312 | V>L | No | gnomAD | |
| rs781321752 | 315 | A>V | No |
ExAC gnomAD |
|
| rs368864621 | 316 | F>L | No |
ESP TOPMed gnomAD |
|
| rs751268082 | 316 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs757098310 | 316 | F>V | No |
ExAC TOPMed gnomAD |
|
| rs763984128 | 318 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1245462924 | 318 | P>S | No | gnomAD | |
| rs1310234570 | 319 | P>S | No | gnomAD | |
| rs1402722094 | 320 | G>A | No |
TOPMed gnomAD |
|
| rs1397577674 | 320 | G>S | No |
TOPMed gnomAD |
|
| rs566480942 | 321 | T>I | No |
1000Genomes ExAC gnomAD |
|
| rs1472220152 | 322 | V>A | No | gnomAD | |
| rs1168709421 | 322 | V>L | No | gnomAD | |
| rs1366364466 | 324 | P>R | No | gnomAD | |
| rs551434705 | 327 | R>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs551434705 | 327 | R>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776871979 | 332 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs761275404 | 334 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1286164384 | 335 | G>S | No | gnomAD | |
| rs1245848725 | 338 | R>C | No | gnomAD | |
| rs1340821507 | 338 | R>H | No | gnomAD | |
| rs1407875814 | 340 | A>T | No | gnomAD | |
| rs967110721 | 343 | L>P | No | TOPMed | |
| rs1042265178 | 345 | A>T | No |
TOPMed gnomAD |
|
| rs1458143967 | 346 | M>V | No | gnomAD | |
| rs1180514338 | 348 | A>P | No | gnomAD | |
| rs1198851115 | 350 | E>D | No | gnomAD | |
| rs749431418 | 354 | P>S | No |
ExAC gnomAD |
|
| rs1292178121 | 356 | G>S | No | gnomAD | |
| rs770738210 | 357 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs142777704 | 358 | Q>H | No |
1000Genomes TOPMed |
|
| rs746970763 | 360 | G>D | No |
ExAC gnomAD |
|
| rs1226356877 | 361 | L>F | No | gnomAD | |
| rs1390088273 | 362 | T>A | No | gnomAD | |
| rs1235451933 | 366 | H>Q | No | TOPMed | |
| rs1328829291 | 366 | H>Y | No | gnomAD | |
| rs148399929 | 367 | F>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1387461280 | 368 | V>M | No | gnomAD | |
| rs200811692 | 370 | A>G | No |
ExAC gnomAD |
|
| rs1157325288 | 370 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM4098334 | 372 | W>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1172481001 | 373 | Q>R | No | gnomAD | |
| rs1248486393 | 374 | A>S | No | gnomAD | |
|
COSM3389743 rs753304095 |
376 | E>K | pancreas [Cosmic] | No |
cosmic curated ExAC gnomAD |
| COSM288372 | 377 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs761024900 | 377 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs750992860 | 378 | S>A | No |
ExAC gnomAD |
|
| COSM6159748 | 378 | S>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs375897463 | 381 | A>T | No |
ESP TOPMed gnomAD |
|
| rs1218529101 | 381 | A>V | No | gnomAD | |
| rs774695747 | 382 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs144459328 | 382 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs763301814 | 383 | C>R | No |
ExAC gnomAD |
|
| rs775648926 | 385 | R>C | No |
ExAC gnomAD |
|
| rs1333958799 | 385 | R>H | No | gnomAD | |
| rs746909694 | 386 | E>D | No |
ExAC gnomAD |
|
| rs769931259 | 386 | E>K | No |
ExAC gnomAD |
|
| TCGA novel | 387 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777835195 | 387 | A>V | No |
ExAC gnomAD |
|
| rs1190725730 | 390 | G>E | No | gnomAD | |
| rs1388440344 | 390 | G>R | No | gnomAD | |
| rs1465534390 | 391 | G>D | No | gnomAD | |
| rs1258609909 | 393 | P>S | No | gnomAD | |
| rs754558450 | 395 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs754558450 | 395 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1340808854 | 396 | T>I | No | gnomAD | |
| rs1211595908 | 396 | T>P | No | gnomAD | |
| rs1340808854 | 396 | T>S | No | gnomAD | |
| rs760761824 | 397 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1362231882 | 399 | T>A | No | gnomAD | |
| rs1261380621 | 403 | S>G | No | TOPMed | |
| rs1446255115 | 403 | S>N | No | gnomAD | |
| rs1600366842 | 403 | S>R | No | Ensembl | |
| rs779523200 | 405 | G>E | No | ExAC | |
| rs1313984335 | 407 | E>K | No | gnomAD | |
| rs1199745801 | 408 | E>K | No | TOPMed | |
| rs1429722527 | 409 | E>V | No | gnomAD | |
| rs1276441146 | 410 | E>K | No | TOPMed | |
| rs750862703 | 412 | E>K | No |
ExAC gnomAD |
|
| rs757730302 | 413 | E>D | No |
ExAC TOPMed |
|
| rs768121518 | 413 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1413367961 | 414 | E>K | No | gnomAD | |
| rs529107942 | 415 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs754367212 | 415 | E>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs529107942 | 415 | E>Q | No |
1000Genomes ExAC gnomAD |
|
| rs1239031263 | 416 | E>D | No | gnomAD | |
| rs1465129765 | 417 | E>K | No | TOPMed | |
| rs191679255 | 419 | E>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs759704025 | 420 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs765228310 | 420 | G>S | No |
ExAC gnomAD |
|
| rs1226292690 | 423 | E>Q | No | gnomAD | |
| rs748080776 | 425 | E>D | No | ExAC | |
| rs772004115 | 425 | E>K | No |
ExAC gnomAD |
|
| rs774073734 | 426 | E>K | No | ExAC | |
| rs1273567897 | 427 | E>D | No | TOPMed | |
| rs1432276673 | 428 | E>D | No | TOPMed | |
| rs1600366625 | 428 | E>G | No | Ensembl | |
| rs768492671 | 429 | G>E | No |
ExAC TOPMed |
|
| rs768492671 | 429 | G>V | No |
ExAC TOPMed |
|
| rs1463870627 | 430 | E>G | No | TOPMed | |
| rs779792835 | 431 | E>A | No |
ExAC TOPMed |
|
| rs779792835 | 431 | E>G | No |
ExAC TOPMed |
|
| rs748930909 | 431 | E>K | No |
ExAC gnomAD |
|
| rs1390246446 | 432 | E>G | No | TOPMed | |
| rs1328003800 | 433 | E>A | No | TOPMed | |
| rs1328003800 | 433 | E>G | No | TOPMed | |
| rs1162911059 | 433 | E>K | No | gnomAD | |
| rs1371827419 | 434 | E>G | No | TOPMed | |
| rs1407344480 | 434 | E>K | No | gnomAD | |
| rs1600366513 | 435 | E>G | No | Ensembl | |
| rs13042058 | 436 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs13042058 | 436 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs780602403 | 436 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed |
| rs13042058 | 436 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs560113368 | 437 | G>E | No | 1000Genomes | |
| rs748211874 | 438 | E>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1600366446 | 438 | E>G | No | Ensembl | |
| rs144899160 | 438 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs748211874 COSM5140684 |
438 | E>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs1249512778 | 439 | G>E | No | gnomAD | |
| rs1184338498 | 441 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1245669689 | 442 | L>* | No |
TOPMed gnomAD |
|
| rs1480653341 | 442 | L>V | No |
TOPMed gnomAD |
|
| rs1245669689 | 442 | L>W | No |
TOPMed gnomAD |
|
| rs1323180072 | 443 | G>R | No | gnomAD | |
| rs1395251585 | 444 | E>G | No | TOPMed | |
| rs1274129101 | 444 | E>K | No | gnomAD | |
| rs752938535 | 447 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs533227646 | 447 | E>K | No | Ensembl | |
| rs1327602524 | 448 | V>E | No |
TOPMed gnomAD |
|
| rs1327602524 | 448 | V>G | No |
TOPMed gnomAD |
|
| rs199992333 | 449 | E>* | No | 1000Genomes | |
| rs759706920 | 450 | E>D | No |
ExAC gnomAD |
|
| rs138025965 | 451 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1472284168 | 451 | E>Q | No | gnomAD | |
| rs1206471984 | 452 | G>V | No | gnomAD | |
| rs761720824 | 453 | D>N | No |
ExAC gnomAD |
|
| rs774335332 | 455 | D>N | No |
ExAC gnomAD |
|
| rs768406399 | 457 | D>G | No | ExAC | |
| rs551419159 | 459 | E>Q | No |
ExAC gnomAD |
|
| rs769450213 | 461 | E>K | No |
ExAC gnomAD |
|
| rs1266004626 | 462 | E>A | No | gnomAD | |
| rs745514430 | 462 | E>K | No | ExAC | |
| rs756885952 | 463 | D>G | No |
ExAC gnomAD |
|
| rs780699266 | 463 | D>N | No |
ExAC gnomAD |
|
| rs747515195 | 464 | E>K | No |
ExAC gnomAD |
|
| COSM3799528 | 465 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs373678745 COSM1200731 |
468 | S>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs373678745 | 468 | S>W | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 469 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1041906979 | 472 | E>V | No | Ensembl | |
| rs1299209466 | 473 | A>G | No | gnomAD | |
| rs1299209466 | 473 | A>V | No | gnomAD | |
| rs1157017690 | 477 | A>S | No | TOPMed | |
| rs945775228 | 477 | A>V | No | TOPMed | |
| rs947501798 | 479 | G>R | No | Ensembl | |
| rs912922404 | 480 | V>A | No | TOPMed | |
| rs1433226896 | 480 | V>L | No |
TOPMed gnomAD |
|
| rs1386115334 | 481 | V>M | No | gnomAD | |
| rs893394617 | 484 | G>D | No | Ensembl | |
| rs760780474 | 484 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs140633295 | 487 | F>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs199738908 | 488 | G>R | No |
1000Genomes TOPMed gnomAD |
|
| rs762912299 | 489 | A>S | No |
ExAC gnomAD |
|
| rs1187517907 | 489 | A>V | No | gnomAD | |
| rs928661373 | 490 | Y>S | No |
TOPMed gnomAD |
|
| rs769493561 | 491 | G>S | No |
ExAC gnomAD |
|
| rs759217770 | 492 | A>S | No |
ExAC gnomAD |
|
| rs1255957529 | 493 | Q>H | No | TOPMed | |
| rs940799141 | 494 | E>Q | No | Ensembl | |
| rs746568860 | 496 | A>V | No |
ExAC gnomAD |
|
| rs748676791 | 497 | Q>R | No |
ExAC gnomAD |
|
| rs1485000639 | 498 | C>W | No | TOPMed | |
| rs748614679 | 500 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs772638786 | 500 | T>S | No |
ExAC gnomAD |
|
| rs149144936 | 503 | F>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1180295262 | 508 | E>K | No | TOPMed | |
| rs1456930774 | 514 | S>G | No |
TOPMed gnomAD |
|
| rs1600365941 | 515 | E>D | No | Ensembl | |
| COSM443807 | 515 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1436558526 | 518 | D>G | No | gnomAD | |
| rs1600365916 | 519 | D>E | No | Ensembl | |
| rs1167453032 | 519 | D>H | No |
TOPMed gnomAD |
|
| rs1167453032 | 519 | D>N | No |
TOPMed gnomAD |
|
| rs1600365904 | 520 | E>D | No | Ensembl | |
| rs566794290 | 522 | E>K | No |
ExAC gnomAD |
|
| rs866979432 | 523 | D>G | No | Ensembl | |
| rs1423568101 | 523 | D>N | No | gnomAD | |
| rs752634687 | 526 | D>G | No |
ExAC gnomAD |
|
| rs1223826972 | 528 | D>N | No | TOPMed | |
|
COSM258766 rs765081320 |
529 | D>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1341635347 | 530 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs141704389 | 532 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs770661500 | 532 | D>N | No |
ExAC gnomAD |
|
|
COSM3363242 rs1600365773 |
533 | E>D | kidney [Cosmic] | No |
cosmic curated Ensembl |
| rs771761547 | 536 | G>S | No |
ExAC gnomAD |
|
| rs779328212 | 539 | V>L | No |
ExAC gnomAD |
|
| rs749565823 | 541 | V>I | No |
ExAC gnomAD |
|
| rs746104845 | 548 | M>I | No |
ExAC gnomAD |
|
| rs1395407026 | 549 | A>S | No | TOPMed | |
| rs1054138522 | 550 | Y>C | No | TOPMed | |
| rs1428909856 | 552 | A>V | No | TOPMed | |
| rs1223829356 | 553 | M>V | No | gnomAD | |
| rs1011315880 | 555 | K>R | No | Ensembl | |
| TCGA novel | 560 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs935629273 | 560 | S>Y | No | TOPMed | |
| rs374655035 | 561 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs200421015 | 562 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1251401829 | 562 | P>S | No | gnomAD | |
| TCGA novel | 563 | I>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1600365629 | 564 | D>A | No | Ensembl | |
| rs1341846872 | 565 | D>E | No | TOPMed | |
| rs893331913 | 565 | D>G | No | Ensembl | |
| rs754967941 | 566 | R>C | No |
ExAC gnomAD |
|
| rs754967941 | 566 | R>G | No |
ExAC gnomAD |
|
| rs753648407 | 566 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs760386998 | 570 | H>N | No |
ExAC gnomAD |
|
| rs773062211 | 572 | L>F | No |
ExAC gnomAD |
|
| rs1407973640 | 573 | H>R | No | gnomAD | |
| TCGA novel | 574 | L>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs557225624 | 576 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs775776155 | 579 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs770202423 | 580 | H>Y | No |
ExAC gnomAD |
|
| rs1469896763 | 581 | V>G | No | TOPMed | |
| rs1411743980 | 581 | V>L | No | gnomAD | |
| rs746086964 | 582 | T>A | No |
ExAC gnomAD |
|
| rs368775626 | 583 | R>G | No |
ESP TOPMed |
|
| rs757341919 | 587 | A>T | No |
ExAC gnomAD |
|
| rs1165926755 | 590 | A>S | No | TOPMed | |
|
COSM3713308 rs747014298 |
590 | A>V | upper_aerodigestive_tract [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs754812338 | 593 | R>* | No |
ExAC gnomAD |
|
| rs753881816 | 593 | R>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 594 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3546275 | 595 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766268613 | 596 | G>A | No |
ExAC gnomAD |
|
| rs1279570028 | 598 | Q>* | No | gnomAD | |
| rs750078348 | 599 | S>N | No | ExAC | |
| rs767027838 | 600 | S>S | No |
ExAC gnomAD |
No associated diseases with P07199
No regional properties for P07199
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P07199 | |||
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromosome | A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information. |
| chromosome, centromeric region | The region of a chromosome that includes the centromeric DNA and associated proteins. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome. |
| condensed chromosome, centromeric region | The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome. |
| nuclear body | Extra-nucleolar nuclear domains usually visualized by confocal microscopy and fluorescent antibodies to specific proteins. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| pericentric heterochromatin | Heterochromatin that is located adjacent to the CENP-A rich centromere 'central core' and characterized by methylated H3 histone at lysine 9 (H3K9me2/H3K9me3). |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| centromeric DNA binding | Binding to a centromere, a region of chromosome where the spindle fibers attach during mitosis and meiosis. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| satellite DNA binding | Binding to satellite DNA, the many tandem repeats (identical or related) of a short basic repeating unit; many have a base composition or other property different from the genome average that allows them to be separated from the bulk (main band) genomic DNA. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
1 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q6B0B8 | TIGD3 | Tigger transposable element-derived protein 3 | Homo sapiens (Human) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGPKRRQLTF | REKSRIIQEV | EENPDLRKGE | IARRFNIPPS | TLSTILKNKR | AILASERKYG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VASTCRKTNK | LSPYDKLEGL | LIAWFQQIRA | AGLPVKGIIL | KEKALRIAEE | LGMDDFTASN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GWLDRFRRRH | GVVSCSGVAR | ARARNAAPRT | PAAPASPAAV | PSEGSGGSTT | GWRAREEQPP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SVAEGYASQD | VFSATETSLW | YDFLPDQAAG | LCGGDGRPRQ | ATQRLSVLLC | ANADGSEKLP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PLVAGKSAKP | RAGQAGLPCD | YTANSKGGVT | TQALAKYLKA | LDTRMAAESR | RVLLLAGRLA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| AQSLDTSGLR | HVQLAFFPPG | TVHPLERGVV | QQVKGHYRQA | MLLKAMAALE | GQDPSGLQLG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LTEALHFVAA | AWQAVEPSDI | AACFREAGFG | GGPNATITTS | LKSEGEEEEE | EEEEEEEEEG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| EGEEEEEEGE | EEEEEGGEGE | ELGEEEEVEE | EGDVDSDEEE | EEDEESSSEG | LEAEDWAQGV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VEAGGSFGAY | GAQEEAQCPT | LHFLEGGEDS | DSDSEEEDDE | EEDDEDEDDD | DDEEDGDEVP |
| 550 | 560 | 570 | 580 | 590 | |
| VPSFGEAMAY | FAMVKRYLTS | FPIDDRVQSH | ILHLEHDLVH | VTRKNHARQA | GVRGLGHQS |