P05532
Gene name |
Kit (Sl) |
Protein name |
Mast/stem cell growth factor receptor Kit |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:16590 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
Descriptions
(Annotation based on sequence homology with P10721)
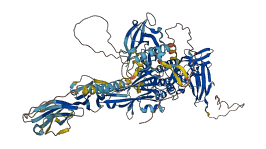
The c-Kit receptor protein-tyrosine kinase is tightly regulated in normal cells, whereas deregulated c-Kit kinase activity is implicated in the pathogenesis of human cancers. Crystal structures of c-Kit kinase reveal a manner in which the small juxtamembrane domain functions to maintain the kinase in an autoinhibited state. Insertion of this autoinhibitory domain into the cleft between the kinase N- and C-lobes flips the DFG motif into its off state, thereby inducing the activation loop to fold back over from its extended conformation in the active kinase where it binds as a pseudosubstrate at the kinase-active center.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
811-836 (Activation loop from InterPro)
Target domain |
592-939 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

2 structures for P05532
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2O26 | X-ray | 250 A | U/W/X/Y | 25-314 | PDB |
| AF-P05532-F1 | Predicted | AlphaFoldDB |
63 variants for P05532
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388759366 | 7 | A>T | No | EVA | |
| rs3388755808 | 7 | A>V | No | EVA | |
| rs217857354 | 40 | H>Q | No | EVA | |
| rs3388766619 | 45 | E>D | No | EVA | |
| rs3388772830 | 48 | V>A | No | EVA | |
| rs3388749478 | 48 | V>F | No | EVA | |
| rs260273061 | 72 | F>S | No | EVA | |
| rs225138717 | 95 | T>A | No | EVA | |
| rs3388766617 | 96 | Y>N | No | EVA | |
| rs245163257 | 101 | S>R | No | EVA | |
| rs223975832 | 122 | G>D | No | EVA | |
| rs238782316 | 123 | L>P | No | EVA | |
| rs3388758922 | 159 | T>M | No | EVA | |
| rs3388766975 | 160 | D>H | No | EVA | |
| rs3388739205 | 161 | L>R | No | EVA | |
| rs33129397 | 169 | A>T | No | EVA | |
| rs3388762771 | 184 | C>S | No | EVA | |
| rs3388772777 | 187 | C>* | No | EVA | |
| 207 | A>E | loss-of-function mutation abolishing ligand binding [UniProt] | No | ||
| rs3388770540 | 218 | P>H | No | EVA | |
| rs3388770540 | 218 | P>L | No | EVA | |
| rs3388759332 | 222 | H>Y | No | EVA | |
| rs3388767030 | 228 | D>N | No | EVA | |
| rs3388762800 | 229 | T>I | No | EVA | |
| rs3388770514 | 231 | T>M | No | EVA | |
| rs3388739235 | 237 | K>* | No | EVA | |
| rs3388758939 | 294 | Y>F | No | EVA | |
| rs3388758959 | 297 | N>I | No | EVA | |
| rs3388759334 | 391 | G>D | No | EVA | |
| rs3388772808 | 396 | L>Q | No | EVA | |
| rs3388767772 | 397 | V>A | No | EVA | |
| rs3388755809 | 406 | V>A | No | EVA | |
| rs247688037 | 425 | I>M | No | EVA | |
| rs3388767805 | 471 | P>Q | No | EVA | |
| rs260250419 | 538 | A>T | No | EVA | |
| rs3395779412 | 545 | V>M | No | EVA | |
| rs3388759306 | 569 | N>I | No | EVA | |
| 586 | E>K | W37 spotting; impaired protein stability and loss of kinase activity [UniProt] | No | ||
| rs3388766143 | 595 | G>V | No | EVA | |
| rs3388770509 | 622 | T>A | No | EVA | |
| rs1131902551 | 657 | V>M | No | EVA | |
| rs1134829284 | 658 | N>K | No | EVA | |
| rs1133113259 | 659 | L>M | No | EVA | |
| rs1134165463 | 662 | A>T | No | EVA | |
| rs1134759679 | 663 | C>F | No | EVA | |
| 664 | T>M | Wv spotting [UniProt] | No | ||
| rs3388763851 | 686 | R>I | No | EVA | |
| rs260462926 | 703 | A>V | No | EVA | |
| rs1131895601 | 709 | L>I | No | EVA | |
| rs233151750 | 713 | T>K | No | EVA | |
| rs3388767023 | 730 | V>I | No | EVA | |
| rs3395388412 | 757 | A>RHLP* | No | EVA | |
| 794 | D>N | W42 spotting; loss of kinase activity and impaired internalization after exposure to KITLG/SCF [UniProt] | No | ||
| rs3388739283 | 801 | L>H | No | EVA | |
| 835 | V>M | W41 spotting; decreased kinase activity [UniProt] | No | ||
| rs3388767804 | 837 | W>C | No | EVA | |
| rs3388767029 | 849 | T>K | No | EVA | |
| rs3388763849 | 862 | W>C | No | EVA | |
| rs3388772756 | 868 | G>R | No | EVA | |
| rs3388762487 | 896 | H>Y | No | EVA | |
| rs3388739208 | 916 | R>G | No | EVA | |
| rs36972615 | 932 | D>E | No | EVA | |
| rs3388755831 | 963 | V>M | No | EVA |
1 associated diseases with P05532
Without disease ID
15 regional properties for P05532
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 592 - 939 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 592 - 925 | IPR001245 |
| conserved_site | Tyrosine-protein kinase, receptor class III, conserved site | 651 - 664 | IPR001824 |
| domain | Immunoglobulin subtype 2 | 49 - 105 | IPR003598-1 |
| domain | Immunoglobulin subtype 2 | 225 - 300 | IPR003598-2 |
| domain | Immunoglobulin subtype 2 | 429 - 501 | IPR003598-3 |
| domain | Immunoglobulin subtype | 43 - 113 | IPR003599-1 |
| domain | Immunoglobulin subtype | 122 - 206 | IPR003599-2 |
| domain | Immunoglobulin subtype | 219 - 311 | IPR003599-3 |
| domain | Immunoglobulin subtype | 323 - 413 | IPR003599-4 |
| domain | Immunoglobulin-like domain | 213 - 311 | IPR007110 |
| active_site | Tyrosine-protein kinase, active site | 790 - 802 | IPR008266 |
| domain | Immunoglobulin | 218 - 307 | IPR013151 |
| binding_site | Protein kinase, ATP binding site | 598 - 626 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 592 - 926 | IPR020635 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| acrosomal vesicle | A structure in the head of a spermatozoon that contains acid hydrolases, and is concerned with the breakdown of the outer membrane of the ovum during fertilization. It lies just beneath the plasma membrane and is derived from the lysosome. |
| cell surface | The external part of the cell wall and/or plasma membrane. |
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic side of plasma membrane | The leaflet the plasma membrane that faces the cytoplasm and any proteins embedded or anchored in it or attached to its surface. |
| external side of plasma membrane | The leaflet of the plasma membrane that faces away from the cytoplasm and any proteins embedded or anchored in it or attached to its surface. |
| extracellular space | That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid. |
| fibrillar center | A structure found most metazoan nucleoli, but not usually found in lower eukaryotes; surrounded by the dense fibrillar component; the zone of transcription from multiple copies of the pre-rRNA genes is in the border region between these two structures. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| cytokine binding | Binding to a cytokine, any of a group of proteins that function to control the survival, growth and differentiation of tissues and cells, and which have autocrine and paracrine activity. |
| metal ion binding | Binding to a metal ion. |
| protease binding | Binding to a protease or a peptidase. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| SH2 domain binding | Binding to a SH2 domain (Src homology 2) of a protein, a protein domain of about 100 amino-acid residues and belonging to the alpha + beta domain class. |
| stem cell factor receptor activity | Combining with stem cell factor (SCF) receptor ligand and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. Stem cell factor is a cytokine that stimulates mast cell growth and differentiation. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. |
80 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton reorganization | A process that is carried out at the cellular level which results in dynamic structural changes to the arrangement of constituent parts of cytoskeletal structures comprising actin filaments and their associated proteins. |
| B cell differentiation | The process in which a precursor cell type acquires the specialized features of a B cell. A B cell is a lymphocyte of B lineage with the phenotype CD19-positive and capable of B cell mediated immunity. |
| cell chemotaxis | The directed movement of a motile cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell population proliferation | The multiplication or reproduction of cells, resulting in the expansion of a cell population. |
| cellular response to thyroid hormone stimulus | A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a thyroid hormone stimulus. |
| chemotaxis | The directed movement of a motile cell or organism, or the directed growth of a cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| cytokine-mediated signaling pathway | The series of molecular signals initiated by the binding of a cytokine to a receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| detection of mechanical stimulus involved in sensory perception of sound | The series of events involved in the perception of sound vibration in which the vibration is received and converted into a molecular signal. |
| developmental pigmentation | The developmental process that results in the deposition of coloring matter in an organism, tissue or cell. |
| digestive tract development | The process whose specific outcome is the progression of the digestive tract over time, from its formation to the mature structure. The digestive tract is the anatomical structure through which food passes and is processed. |
| ectopic germ cell programmed cell death | Programmed cell death of an errant germ line cell that is outside the normal migratory path or ectopic to the gonad. This is an important mechanism of regulating germ cell survival within the embryo. |
| embryonic hemopoiesis | The stages of blood cell formation that take place within the embryo. |
| epithelial cell proliferation | The multiplication or reproduction of epithelial cells, resulting in the expansion of a cell population. Epithelial cells make up the epithelium, the covering of internal and external surfaces of the body, including the lining of vessels and other small cavities. It consists of cells joined by small amounts of cementing substances. |
| erythrocyte differentiation | The process in which a myeloid precursor cell acquires specializes features of an erythrocyte. |
| erythropoietin-mediated signaling pathway | The series of molecular signals initiated by erythropoietin (EPO) binding to the erythropoietin receptor (EPO-R) on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| Fc receptor signaling pathway | The series of molecular signals initiated by the binding of the Fc portion of an immunoglobulin to an Fc receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. The Fc portion of an immunoglobulin is its C-terminal constant region. |
| germ cell development | The process whose specific outcome is the progression of an immature germ cell over time, from its formation to the mature structure (gamete). A germ cell is any reproductive cell in a multicellular organism. |
| germ cell migration | The orderly movement of a cell specialized to produce haploid gametes through the embryo from its site of production to the place where the gonads will form. |
| glycosphingolipid metabolic process | The chemical reactions and pathways involving glycosphingolipids, any compound with residues of sphingoid and at least one monosaccharide. |
| hematopoietic progenitor cell differentiation | The process in which precursor cell type acquires the specialized features of a hematopoietic progenitor cell, a class of cell types including myeloid progenitor cells and lymphoid progenitor cells. |
| hematopoietic stem cell migration | The orderly movement of a hematopoietic stem cell from one site to another. A hematopoietic stem cell is a cell from which all cells of the lymphoid and myeloid lineages develop, including blood cells and cells of the immune system. |
| hemopoiesis | The process whose specific outcome is the progression of the myeloid and lymphoid derived organ/tissue systems of the blood and other parts of the body over time, from formation to the mature structure. The site of hemopoiesis is variable during development, but occurs primarily in bone marrow or kidney in many adult vertebrates. |
| immature B cell differentiation | The process in which a precursor cell type acquires the specialized features of an immature B cell. |
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| Kit signaling pathway | The series of molecular signals initiated by the binding of stem cell factor to the tyrosine kinase receptor KIT on the surface of a target cell, and ending with regulation of a downstream cellular process, e.g. transcription. Stem cell factor (KIT ligand) binding to the receptor Kit mediates receptor dimerization, activation of its intrinsic tyrosine kinase activity and autophosphorylation. The activated receptor then phosphorylates various substrates, thereby activating distinct signaling cascades within the cell that trigger a change in state or activity of the cell. |
| lamellipodium assembly | Formation of a lamellipodium, a thin sheetlike extension of the surface of a migrating cell. |
| lymphoid progenitor cell differentiation | The process in which a precursor cell type acquires the specialized features of a lymphoid progenitor cell. Lymphoid progenitor cells include progenitor cells for any of the lymphoid lineages. |
| male gonad development | The process whose specific outcome is the progression of the male gonad over time, from its formation to the mature structure. |
| mast cell chemotaxis | The movement of a mast cell in response to an external stimulus. |
| mast cell degranulation | The regulated exocytosis of secretory granules containing preformed mediators such as histamine, serotonin, and neutral proteases by a mast cell. |
| mast cell differentiation | The process in which a relatively unspecialized myeloid precursor cell acquires the specialized features of a mast cell. A mast cell is a cell that is found in almost all tissues containing numerous basophilic granules and capable of releasing large amounts of histamine and heparin upon activation. |
| mast cell proliferation | The expansion of a mast cell population by cell division. |
| megakaryocyte development | The process whose specific outcome is the progression of a megakaryocyte cell over time, from its formation to the mature structure. Megakaryocyte development does not include the steps involved in committing a cell to a megakaryocyte fate. A megakaryocyte is a giant cell 50 to 100 micron in diameter, with a greatly lobulated nucleus, found in the bone marrow. |
| melanocyte adhesion | The attachment of a melanocyte to another cell via adhesion molecules. |
| melanocyte differentiation | The process in which a relatively unspecialized cell acquires specialized features of a melanocyte. |
| melanocyte migration | The orderly movement of melanocytes from one site to another, often during the development of a multicellular organism. A melanocyte is a pigment cell derived from the neural crest. It contains melanin-filled pigment granules, which give a brown to black appearance. |
| myeloid progenitor cell differentiation | The process in which a precursor cell type acquires the specialized features of a myeloid progenitor cell. Myeloid progenitor cells include progenitor cells for any of the myeloid lineages. |
| negative regulation of developmental process | Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult). |
| negative regulation of programmed cell death | Any process that stops, prevents, or reduces the frequency, rate or extent of programmed cell death, cell death resulting from activation of endogenous cellular processes. |
| negative regulation of reproductive process | Any process that stops, prevents, or reduces the frequency, rate or extent of reproductive process. |
| ovarian follicle development | The process whose specific outcome is the progression of the ovarian follicle over time, from its formation to the mature structure. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| pigmentation | The accumulation of pigment in an organism, tissue or cell, either by increased deposition or by increased number of cells. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of colon smooth muscle contraction | Any process that activates or increases the frequency, rate or extent of colon smooth muscle contraction. |
| positive regulation of dendritic cell cytokine production | Any process that activates or increases the frequency, rate, or extent of dendritic cell cytokine production. |
| positive regulation of DNA-binding transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of kinase activity | Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| positive regulation of long-term neuronal synaptic plasticity | A process that increases long-term neuronal synaptic plasticity, the ability of neuronal synapses to change long-term as circumstances require. Long-term neuronal synaptic plasticity generally involves increase or decrease in actual synapse numbers. |
| positive regulation of MAP kinase activity | Any process that activates or increases the frequency, rate or extent of MAP kinase activity. |
| positive regulation of MAPK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade. |
| positive regulation of mast cell cytokine production | Any process that activates or increases the frequency, rate, or extent of mast cell cytokine production. |
| positive regulation of mast cell proliferation | Any process that activates or increases the rate or extent of mast cell proliferation. |
| positive regulation of Notch signaling pathway | Any process that activates or increases the frequency, rate or extent of the Notch signaling pathway. |
| positive regulation of pseudopodium assembly | Any process that activates or increases the frequency, rate or extent of the assembly of pseudopodia. |
| positive regulation of pyloric antrum smooth muscle contraction | Any process that increases the frequency, rate or extent of any pyloric antrum smooth muscle contraction. |
| positive regulation of receptor signaling pathway via JAK-STAT | Any process that activates or increases the frequency, rate or extent of the JAK-STAT signaling pathway activity. |
| positive regulation of small intestine smooth muscle contraction | Any process that activates or increases the frequency, rate or extent of small intestine smooth muscle contraction. |
| positive regulation of tyrosine phosphorylation of STAT protein | Any process that activates or increases the frequency, rate or extent of the introduction of a phosphate group to a tyrosine residue of a STAT (Signal Transducer and Activator of Transcription) protein. |
| positive regulation of vascular associated smooth muscle cell differentiation | Any process that activates or increases the frequency, rate or extent of vascular smooth muscle cell differentiation. |
| programmed cell death | A process which begins when a cell receives an internal or external signal and activates a series of biochemical events (signaling pathway). The process ends with the death of the cell. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of bile acid metabolic process | Any process that modulates the frequency, rate or extent of bile acid metabolic process. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| response to cadmium ion | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cadmium (Cd) ion stimulus. |
| response to radiation | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an electromagnetic radiation stimulus. Electromagnetic radiation is a propagating wave in space with electric and magnetic components. These components oscillate at right angles to each other and to the direction of propagation. |
| somatic stem cell division | The self-renewing division of a somatic stem cell, a stem cell that can give rise to cell types of the body other than those of the germ-line. |
| somatic stem cell population maintenance | Any process by which an organism retains a population of somatic stem cells, undifferentiated cells in the embryo or adult which can undergo unlimited division and give rise to cell types of the body other than those of the germ-line. |
| spermatid development | The process whose specific outcome is the progression of a spermatid over time, from its formation to the mature structure. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
| stem cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a stem cell. A stem cell is a cell that retains the ability to divide and proliferate throughout life to provide progenitor cells that can differentiate into specialized cells. |
| T cell differentiation | The process in which a precursor cell type acquires characteristics of a more mature T-cell. A T cell is a type of lymphocyte whose definin characteristic is the expression of a T cell receptor complex. |
| tongue development | The process whose specific outcome is the progression of the tongue over time, from its formation to the mature structure. The tongue is the movable, muscular organ on the floor of the mouth of most vertebrates, in many other mammals is the principal organ of taste, aids in the prehension of food, in swallowing, and in modifying the voice as in speech. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| visual learning | Any process in an organism in which a change in behavior of an individual occurs in response to repeated exposure to a visual cue. |
102 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q06805 | TIE1 | Tyrosine-protein kinase receptor Tie-1 | Bos taurus (Bovine) | PR |
| Q06807 | TEK | Angiopoietin-1 receptor | Bos taurus (Bovine) | SS |
| P43481 | KIT | Mast/stem cell growth factor receptor Kit | Bos taurus (Bovine) | SS |
| P13369 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Felis catus (Cat) (Felis silvestris catus) | SS |
| Q28889 | KIT | Mast/stem cell growth factor receptor Kit | Felis catus (Cat) (Felis silvestris catus) | SS |
| P18460 | FGFR3 | Fibroblast growth factor receptor 3 | Gallus gallus (Chicken) | SS |
| P21804 | FGFR1 | Fibroblast growth factor receptor 1 | Gallus gallus (Chicken) | SS |
| Q9PUF6 | PDGFRA | Platelet-derived growth factor receptor alpha | Gallus gallus (Chicken) | SS |
| Q8QHL3 | FLT1 | Vascular endothelial growth factor receptor 1 | Gallus gallus (Chicken) | SS |
| P18461 | FGFR2 | Fibroblast growth factor receptor 2 | Gallus gallus (Chicken) | SS |
| Q08156 | KIT | Mast/stem cell growth factor receptor Kit | Gallus gallus (Chicken) | SS |
| O97799 | KIT | Mast/stem cell growth factor receptor Kit | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| Q07407 | htl | Fibroblast growth factor receptor homolog 1 | Drosophila melanogaster (Fruit fly) | PR |
| Q6J9G0 | STYK1 | Tyrosine-protein kinase STYK1 | Homo sapiens (Human) | PR |
| P36888 | FLT3 | Receptor-type tyrosine-protein kinase FLT3 | Homo sapiens (Human) | EV |
| P16234 | PDGFRA | Platelet-derived growth factor receptor alpha | Homo sapiens (Human) | EV |
| P09619 | PDGFRB | Platelet-derived growth factor receptor beta | Homo sapiens (Human) | EV |
| P35916 | FLT4 | Vascular endothelial growth factor receptor 3 | Homo sapiens (Human) | SS |
| P35968 | KDR | Vascular endothelial growth factor receptor 2 | Homo sapiens (Human) | EV |
| P17948 | FLT1 | Vascular endothelial growth factor receptor 1 | Homo sapiens (Human) | SS |
| P07333 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Homo sapiens (Human) | SS |
| P22455 | FGFR4 | Fibroblast growth factor receptor 4 | Homo sapiens (Human) | SS |
| P22607 | FGFR3 | Fibroblast growth factor receptor 3 | Homo sapiens (Human) | EV |
| P07949 | RET | Proto-oncogene tyrosine-protein kinase receptor Ret | Homo sapiens (Human) | EV |
| P35590 | TIE1 | Tyrosine-protein kinase receptor Tie-1 | Homo sapiens (Human) | PR |
| Q02763 | TEK | Angiopoietin-1 receptor | Homo sapiens (Human) | SS |
| P11362 | FGFR1 | Fibroblast growth factor receptor 1 | Homo sapiens (Human) | EV |
| P21802 | FGFR2 | Fibroblast growth factor receptor 2 | Homo sapiens (Human) | EV |
| P10721 | KIT | Mast/stem cell growth factor receptor Kit | Homo sapiens (Human) | EV |
| Q91V87 | Fgfrl1 | Fibroblast growth factor receptor-like 1 | Mus musculus (Mouse) | PR |
| Q6J9G1 | Styk1 | Tyrosine-protein kinase STYK1 | Mus musculus (Mouse) | PR |
| P35546 | Ret | Proto-oncogene tyrosine-protein kinase receptor Ret | Mus musculus (Mouse) | SS |
| Q61006 | Musk | Muscle, skeletal receptor tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q9Z138 | Ror2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Mus musculus (Mouse) | SS |
| Q9Z139 | Ror1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Mus musculus (Mouse) | PR |
| Q62371 | Ddr2 | Discoidin domain-containing receptor 2 | Mus musculus (Mouse) | SS |
| Q03146 | Ddr1 | Epithelial discoidin domain-containing receptor 1 | Mus musculus (Mouse) | SS |
| P15209 | Ntrk2 | BDNF/NT-3 growth factors receptor | Mus musculus (Mouse) | SS |
| Q3UFB7 | Ntrk1 | High affinity nerve growth factor receptor | Mus musculus (Mouse) | SS |
| Q6VNS1 | Ntrk3 | NT-3 growth factor receptor | Mus musculus (Mouse) | SS |
| P35917 | Flt4 | Vascular endothelial growth factor receptor 3 | Mus musculus (Mouse) | SS |
| P35969 | Flt1 | Vascular endothelial growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P35918 | Kdr | Vascular endothelial growth factor receptor 2 | Mus musculus (Mouse) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| P97793 | Alk | ALK tyrosine kinase receptor | Mus musculus (Mouse) | SS |
| Q9WTL4 | Insrr | Insulin receptor-related protein | Mus musculus (Mouse) | SS |
| Q60751 | Igf1r | Insulin-like growth factor 1 receptor | Mus musculus (Mouse) | SS |
| P15208 | Insr | Insulin receptor | Mus musculus (Mouse) | SS |
| P16092 | Fgfr1 | Fibroblast growth factor receptor 1 | Mus musculus (Mouse) | SS |
| P21803 | Fgfr2 | Fibroblast growth factor receptor 2 | Mus musculus (Mouse) | SS |
| Q03142 | Fgfr4 | Fibroblast growth factor receptor 4 | Mus musculus (Mouse) | PR |
| Q60805 | Mertk | Tyrosine-protein kinase Mer | Mus musculus (Mouse) | SS |
| Q62190 | Mst1r | Macrophage-stimulating protein receptor | Mus musculus (Mouse) | SS |
| Q00993 | Axl | Tyrosine-protein kinase receptor UFO | Mus musculus (Mouse) | PR |
| P55144 | Tyro3 | Tyrosine-protein kinase receptor TYRO3 | Mus musculus (Mouse) | SS |
| P70424 | Erbb2 | Receptor tyrosine-protein kinase erbB-2 | Mus musculus (Mouse) | SS |
| Q61527 | Erbb4 | Receptor tyrosine-protein kinase erbB-4 | Mus musculus (Mouse) | SS |
| Q61526 | Erbb3 | Receptor tyrosine-protein kinase erbB-3 | Mus musculus (Mouse) | SS |
| Q01279 | Egfr | Epidermal growth factor receptor | Mus musculus (Mouse) | SS |
| Q01887 | Ryk | Tyrosine-protein kinase RYK | Mus musculus (Mouse) | PR |
| Q02858 | Tek | Angiopoietin-1 receptor | Mus musculus (Mouse) | SS |
| Q06806 | Tie1 | Tyrosine-protein kinase receptor Tie-1 | Mus musculus (Mouse) | SS |
| P05622 | Pdgfrb | Platelet-derived growth factor receptor beta | Mus musculus (Mouse) | SS |
| P09581 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Mus musculus (Mouse) | SS |
| P26618 | Pdgfra | Platelet-derived growth factor receptor alpha | Mus musculus (Mouse) | SS |
| Q00342 | Flt3 | Receptor-type tyrosine-protein kinase FLT3 | Mus musculus (Mouse) | SS |
| Q61851 | Fgfr3 | Fibroblast growth factor receptor 3 | Mus musculus (Mouse) | PR |
| P16056 | Met | Hepatocyte growth factor receptor | Mus musculus (Mouse) | PR |
| Q2HWD6 | KIT | Mast/stem cell growth factor receptor Kit | Sus scrofa (Pig) | SS |
| Q7TQM3 | Fgfrl1 | Fibroblast growth factor receptor-like 1 | Rattus norvegicus (Rat) | PR |
| P53767 | Flt1 | Vascular endothelial growth factor receptor 1 | Rattus norvegicus (Rat) | PR |
| P20786 | Pdgfra | Platelet-derived growth factor receptor alpha | Rattus norvegicus (Rat) | SS |
| Q91ZT1 | Flt4 | Vascular endothelial growth factor receptor 3 | Rattus norvegicus (Rat) | SS |
| Q04589 | Fgfr1 | Fibroblast growth factor receptor 1 | Rattus norvegicus (Rat) | SS |
| G3V9H8 | Ret | Proto-oncogene tyrosine-protein kinase receptor Ret | Rattus norvegicus (Rat) | SS |
| Q498D6 | Fgfr4 | Fibroblast growth factor receptor 4 | Rattus norvegicus (Rat) | PR |
| Q05030 | Pdgfrb | Platelet-derived growth factor receptor beta | Rattus norvegicus (Rat) | SS |
| O08775 | Kdr | Vascular endothelial growth factor receptor 2 | Rattus norvegicus (Rat) | SS |
| Q17833 | old-1 | Tyrosine-protein kinase receptor old-1 | Caenorhabditis elegans | PR |
| Q19238 | F09A5.2 | Putative tyrosine-protein kinase F09A5.2 | Caenorhabditis elegans | SS |
| Q10656 | egl-15 | Myoblast growth factor receptor egl-15 | Caenorhabditis elegans | PR |
| P34892 | kin-16 | Receptor-like tyrosine-protein kinase kin-16 | Caenorhabditis elegans | PR |
| G5ED65 | ver-1 | Protein ver-1 | Caenorhabditis elegans | PR |
| Q9S9M2 | WAKL4 | Wall-associated receptor kinase-like 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7X8C5 | WAKL2 | Wall-associated receptor kinase-like 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VYA3 | WAKL10 | Wall-associated receptor kinase-like 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C9L5 | WAKL9 | Wall-associated receptor kinase-like 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M092 | WAKL17 | Wall-associated receptor kinase-like 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8AXB3 | kdrl | Vascular endothelial growth factor receptor kdr-like | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q5GIT4 | kdr | Vascular endothelial growth factor receptor 2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O73791 | tek | Angiopoietin-1 receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q90Z00 | fgfr1a | Fibroblast growth factor receptor 1-A | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q8JG38 | fgfr2 | Fibroblast growth factor receptor 2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9I8N6 | csf1r | Macrophage colony-stimulating factor 1 receptor | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q90413 | fgfr4 | Fibroblast growth factor receptor 4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q9DE49 | pdgfra | Platelet-derived growth factor receptor alpha | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q5MD89 | flt4 | Vascular endothelial growth factor receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q8JFR5 | kita | Mast/stem cell growth factor receptor kita | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRGARGAWDL | LCVLLVLLRG | QTATSQPSAS | PGEPSPPSIH | PAQSELIVEA | GDTLSLTCID |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PDFVRWTFKT | YFNEMVENKK | NEWIQEKAEA | TRTGTYTCSN | SNGLTSSIYV | FVRDPAKLFL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VGLPLFGKED | SDALVRCPLT | DPQVSNYSLI | ECDGKSLPTD | LTFVPNPKAG | ITIKNVKRAY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| HRLCVRCAAQ | RDGTWLHSDK | FTLKVRAAIK | AIPVVSVPET | SHLLKKGDTF | TVVCTIKDVS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TSVNSMWLKM | NPQPQHIAQV | KHNSWHRGDF | NYERQETLTI | SSARVDDSGV | FMCYANNTFG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SANVTTTLKV | VEKGFINISP | VKNTTVFVTD | GENVDLVVEY | EAYPKPEHQQ | WIYMNRTSAN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KGKDYVKSDN | KSNIRYVNQL | RLTRLKGTEG | GTYTFLVSNS | DASASVTFNV | YVNTKPEILT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| YDRLINGMLQ | CVAEGFPEPT | IDWYFCTGAE | QRCTTPVSPV | DVQVQNVSVS | PFGKLVVQSS |
| 490 | 500 | 510 | 520 | 530 | 540 |
| IDSSVFRHNG | TVECKASNDV | GKSSAFFNFA | FKGNNKEQIQ | AHTLFTPLLI | GFVVAAGAMG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| IIVMVLTYKY | LQKPMYEVQW | KVVEEINGNN | YVYIDPTQLP | YDHKWEFPRN | RLSFGKTLGA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GAFGKVVEAT | AYGLIKSDAA | MTVAVKMLKP | SAHLTEREAL | MSELKVLSYL | GNHMNIVNLL |
| 670 | 680 | 690 | 700 | 710 | 720 |
| GACTVGGPTL | VITEYCCYGD | LLNFLRRKRD | SFIFSKQEEQ | AEAALYKNLL | HSTEPSCDSS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| NEYMDMKPGV | SYVVPTKTDK | RRSARIDSYI | ERDVTPAIME | DDELALDLDD | LLSFSYQVAK |
| 790 | 800 | 810 | 820 | 830 | 840 |
| GMAFLASKNC | IHRDLAARNI | LLTHGRITKI | CDFGLARDIR | NDSNYVVKGN | ARLPVKWMAP |
| 850 | 860 | 870 | 880 | 890 | 900 |
| ESIFSCVYTF | ESDVWSYGIF | LWELFSLGSS | PYPGMPVDSK | FYKMIKEGFR | MVSPEHAPAE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| MYDVMKTCWD | ADPLKRPTFK | QVVQLIEKQI | SDSTKHIYSN | LANCNPNPEN | PVVVDHSVRV |
| 970 | |||||
| NSVGSSASST | QPLLVHEDA |