P04896
Gene name |
GNAS (GNAS1) |
Protein name |
Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
Names |
Adenylate cyclase-stimulating G alpha protein |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:281793 |
EC number |
|
Protein Class |
|
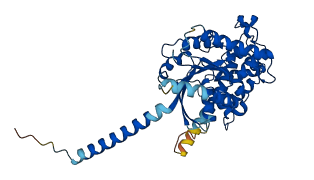
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

43 structures for P04896
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1AZS | X-ray | 230 A | C | 1-394 | PDB |
| 1AZT | X-ray | 230 A | A/B | 1-394 | PDB |
| 1CJK | X-ray | 300 A | C | 1-394 | PDB |
| 1CJT | X-ray | 280 A | C | 1-394 | PDB |
| 1CJU | X-ray | 280 A | C | 1-394 | PDB |
| 1CJV | X-ray | 300 A | C | 1-394 | PDB |
| 1CS4 | X-ray | 250 A | C | 1-394 | PDB |
| 1CUL | X-ray | 240 A | C | 1-394 | PDB |
| 1TL7 | X-ray | 280 A | C | 1-394 | PDB |
| 1U0H | X-ray | 290 A | C | 1-394 | PDB |
| 2GVD | X-ray | 290 A | C | 1-394 | PDB |
| 2GVZ | X-ray | 327 A | C | 1-394 | PDB |
| 3C14 | X-ray | 268 A | C | 1-394 | PDB |
| 3C15 | X-ray | 278 A | C | 1-394 | PDB |
| 3C16 | X-ray | 287 A | C | 1-394 | PDB |
| 3G82 | X-ray | 311 A | C | 1-394 | PDB |
| 3MAA | X-ray | 300 A | C | 1-394 | PDB |
| 3SN6 | X-ray | 320 A | A | 1-394 | PDB |
| 6NBF | EM | 300 A | PDB | ||
| 6NBH | EM | 350 A | PDB | ||
| 6NBI | EM | 400 A | PDB | ||
| 6R3Q | EM | 340 A | B | 1-394 | PDB |
| 6R4O | EM | 420 A | B | 1-394 | PDB |
| 6R4P | EM | 310 A | B | 1-394 | PDB |
| 7D68 | EM | 300 A | PDB | ||
| 7DTY | EM | 298 A | A | 1-394 | PDB |
| 7EQ1 | EM | 330 A | PDB | ||
| 7FIM | EM | 340 A | A | 1-394 | PDB |
| 7FIN | EM | 310 A | A | 1-394 | PDB |
| 7FIY | EM | 340 A | A | 1-394 | PDB |
| 7JJO | EM | 260 A | A | 1-394 | PDB |
| 7PDE | EM | 450 A | B | 1-394 | PDB |
| 7PDF | EM | 380 A | B | 1-394 | PDB |
| 7S0G | EM | 386 A | A | 384-394 | PDB |
| 7VQX | EM | 274 A | A | 1-394 | PDB |
| 7WBJ | EM | 342 A | A | 1-394 | PDB |
| 8BUZ | EM | 350 A | B | 1-394 | PDB |
| 8BV5 | EM | 354 A | B | 1-394 | PDB |
| 8DCR | EM | 260 A | A | 1-394 | PDB |
| 8DCS | EM | 250 A | A | 1-394 | PDB |
| 8ITL | EM | 323 A | A | 1-394 | PDB |
| 8ITM | EM | 313 A | A | 1-394 | PDB |
| AF-P04896-F1 | Predicted | AlphaFoldDB |
48 variants for P04896
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs457063131 | 55 | T>I | No | EVA | |
| rs435261957 | 60 | M>I | No | EVA | |
| rs470531739 | 68 | F>L | No | EVA | |
| rs452801441 | 71 | E>V | No | EVA | |
| rs447406450 | 76 | D>G | No | EVA | |
| rs477047276 | 85 | D>N | No | EVA | |
| rs482345368 | 105 | T>P | No | EVA | |
| rs469162113 | 144 | P>H | No | EVA | |
| rs472786815 | 146 | F>C | No | EVA | |
| rs443085661 | 146 | F>I | No | EVA | |
| rs472786815 | 146 | F>S | No | EVA | |
| rs466503374 | 221 | M>I | No | EVA | |
| rs433610251 | 221 | M>R | No | EVA | |
| rs433610251 | 221 | M>T | No | EVA | |
| rs455419465 | 221 | M>V | No | EVA | |
| rs445558785 | 222 | F>Y | No | EVA | |
| rs478513568 | 223 | D>G | No | EVA | |
| rs469621564 | 224 | V>E | No | EVA | |
| rs450944270 | 226 | G>V | No | EVA | |
| rs480528916 | 227 | Q>E | No | EVA | |
| rs462121041 | 229 | D>N | No | EVA | |
| rs440444600 | 230 | E>D | No | EVA | |
| rs479653384 | 233 | K>Q | No | EVA | |
| rs458035050 | 238 | F>L | No | EVA | |
| rs438708256 | 239 | N>S | No | EVA | |
| rs110996434 | 261 | N>H | No | EVA | |
| rs109049760 | 261 | N>T | No | EVA | |
| rs473990741 | 267 | Q>L | No | EVA | |
| rs455454369 | 270 | L>P | No | EVA | |
| rs474104728 | 272 | L>R | No | EVA | |
| rs439547580 | 284 | T>P | No | EVA | |
| rs482262566 | 289 | L>V | No | EVA | |
| rs463606441 | 290 | F>V | No | EVA | |
| rs462857894 | 291 | L>F | No | EVA | |
| rs461904231 | 295 | D>E | No | EVA | |
| rs474025418 | 295 | D>V | No | EVA | |
| rs440153407 | 307 | K>* | No | EVA | |
| rs473026823 | 309 | E>A | No | EVA | |
| rs451291357 | 314 | E>K | No | EVA | |
| rs432816092 | 315 | F>I | No | EVA | |
| rs457512975 | 321 | P>H | No | EVA | |
| rs434887362 | 322 | E>D | No | EVA | |
| rs468630669 | 322 | E>K | No | EVA | |
| rs446957592 | 322 | E>V | No | EVA | |
| rs436731237 | 359 | C>F | No | EVA | |
| rs436731237 | 359 | C>Y | No | EVA | |
| rs463194544 | 381 | D>E | No | EVA | |
| rs437363707 | 384 | Q>H | No | EVA |
No associated diseases with P04896
No regional properties for P04896
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P04896 | |||
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| heterotrimeric G-protein complex | Any of a family of heterotrimeric GTP-binding and hydrolyzing proteins; they belong to a superfamily of GTPases that includes monomeric proteins such as EF-Tu and RAS. Heterotrimeric G-proteins consist of three subunits; the alpha subunit contains the guanine nucleotide binding site and possesses GTPase activity; the beta and gamma subunits are tightly associated and function as a beta-gamma heterodimer; extrinsic plasma membrane proteins (cytoplasmic face) that function as a complex to transduce signals from G protein-coupled receptors to an effector protein. |
| intrinsic component of membrane | The component of a membrane consisting of the gene products having some covalently attached portion, for example part of a peptide sequence or some other covalently attached group such as a GPI anchor, which spans or is embedded in one or both leaflets of the membrane. |
| trans-Golgi network membrane | The lipid bilayer surrounding any of the compartments that make up the trans-Golgi network. |
12 GO annotations of molecular function
| Name | Definition |
|---|---|
| adenylate cyclase activator activity | Increases the activity of the enzyme that catalyzes the reaction: ATP = 3',5'-cyclic AMP + diphosphate. |
| beta-2 adrenergic receptor binding | Binding to a beta-2 adrenergic receptor. |
| corticotropin-releasing hormone receptor 1 binding | Binding to a corticotropin-releasing hormone receptor 1 (CRHR1). CRHR1 is the major subtype in the pituitary corticotroph, and mediates the stimulatory actions of corticotropin-releasing hormone on corticotropin hormone secretion. CRHR1 are also located in cortical areas of the brain, cerebellum and limbic system. |
| D1 dopamine receptor binding | Binding to a D1 dopamine receptor. |
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| insulin-like growth factor receptor binding | Binding to an insulin-like growth factor receptor. |
| ionotropic glutamate receptor binding | Binding to an ionotropic glutamate receptor. Ionotropic glutamate receptors bind glutamate and exert an effect through the regulation of ion channels. |
| metal ion binding | Binding to a metal ion. |
| mu-type opioid receptor binding | Binding to a mu-type opioid receptor. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| adenylate cyclase-activating adrenergic receptor signaling pathway | An adenylate cyclase-activating G protein-coupled receptor signaling pathway initiated by a ligand binding to an adrenergic receptor on the surface of the target cell, and ending with the regulation of a downstream cellular process. |
| adenylate cyclase-activating dopamine receptor signaling pathway | An adenylate cyclase-activating G protein-coupled receptor signaling pathway initiated by dopamine binding to its receptor, and ending with the regulation of a downstream cellular process. |
| adenylate cyclase-activating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation of adenylyl cyclase activity and a subsequent increase in the intracellular concentration of cyclic AMP (cAMP). |
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation or inhibition of adenylyl cyclase activity and a subsequent change in the intracellular concentration of cyclic AMP (cAMP). |
| bone development | The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components. |
| cognition | The operation of the mind by which an organism becomes aware of objects of thought or perception; it includes the mental activities associated with thinking, learning, and memory. |
| developmental growth | The increase in size or mass of an entire organism, a part of an organism or a cell, where the increase in size or mass has the specific outcome of the progression of the organism over time from one condition to another. |
| hair follicle placode formation | The developmental process in which a hair placode forms. An hair follicle placode is a thickening of the ectoderm that will give rise to the hair follicle bud. |
| platelet aggregation | The adhesion of one platelet to one or more other platelets via adhesion molecules. |
| positive regulation of cAMP-mediated signaling | Any process which activates, maintains or increases the frequency, rate or extent of cAMP-mediated signaling. |
| positive regulation of GTPase activity | Any process that activates or increases the activity of a GTPase. |
| sensory perception of chemical stimulus | The series of events required for an organism to receive a sensory chemical stimulus, convert it to a molecular signal, and recognize and characterize the signal. This is a neurological process. |
18 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P38408 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Bos taurus (Bovine) | PR |
| P0C7Q4 | GNAT3 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04696 | GNAT2 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04695 | GNAT1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P63097 | GNAI1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P08239 | GNAO1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P63091 | GNAS | Guanine nucleotide-binding protein G(s) subunit alpha | Canis lupus familiaris (Dog) (Canis familiaris) | PR |
| P38405 | GNAL | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P29797 | GNAS | Guanine nucleotide-binding protein G(s) subunit alpha | Sus scrofa (Pig) | PR |
| P93564 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum tuberosum (Potato) | SS |
| P38406 | Gnal | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| Q63803 | Gnas | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| Q0DJ33 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Oryza sativa subsp. japonica (Rice) | SS |
| P49084 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| P93163 | GPA2 | Guanine nucleotide-binding protein alpha-2 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| O80462 | XLG1 | Extra-large guanine nucleotide-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P18064 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P26981 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGCLGNSKTE | DQRNEEKAQR | EANKKIEKQL | QKDKQVYRAT | HRLLLLGAGE | SGKSTIVKQM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RILHVNGFNG | EGGEEDPQAA | RSNSDGEKAT | KVQDIKNNLK | EAIETIVAAM | SNLVPPVELA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NPENQFRVDY | ILSVMNVPDF | DFPPEFYEHA | KALWEDEGVR | ACYERSNEYQ | LIDCAQYFLD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KIDVIKQDDY | VPSDQDLLRC | RVLTSGIFET | KFQVDKVNFH | MFDVGGQRDE | RRKWIQCFND |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VTAIIFVVAS | SSYNMVIRED | NQTNRLQEAL | NLFKSIWNNR | WLRTISVILF | LNKQDLLAEK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VLAGKSKIED | YFPEFARYTT | PEDATPEPGE | DPRVTRAKYF | IRDEFLRIST | ASGDGRHYCY |
| 370 | 380 | 390 | |||
| PHFTCAVDTE | NIRRVFNDCR | DIIQRMHLRQ | YELL |