P04696
Gene name |
GNAT2 |
Protein name |
Guanine nucleotide-binding protein G |
Names |
t subunit alpha-2 , Transducin alpha-2 chain |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:281795 |
EC number |
|
Protein Class |
|
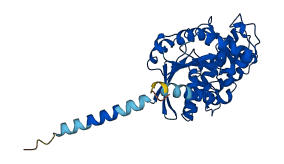
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
184-348 (Ras-like domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Goricanec D et al. (2016) "Conformational dynamics of a G-protein α subunit is tightly regulated by nucleotide binding", Proceedings of the National Academy of Sciences of the United States of America, 113, E3629-38
- Coleman DE et al. (1999) "Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex", The Journal of biological chemistry, 274, 16669-72
- Lutz S et al. (2007) "Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs", Science (New York, N.Y.), 318, 1923-7
Autoinhibited structure

Activated structure

3 structures for P04696
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6PGS | X-ray | 290 A | B | 344-354 | PDB |
| 6PH7 | X-ray | 290 A | B | 344-354 | PDB |
| AF-P04696-F1 | Predicted | AlphaFoldDB |
40 variants for P04696
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs472853112 | 24 | Q>P | No | EVA | |
| rs441436892 | 26 | D>E | No | EVA | |
| rs471329908 | 27 | A>D | No | EVA | |
| rs458144472 | 27 | A>T | No | EVA | |
| rs463192408 | 29 | K>N | No | EVA | |
| rs443235507 | 29 | K>T | No | EVA | |
| rs480058632 | 30 | E>D | No | EVA | |
| rs448781104 | 31 | A>S | No | EVA | |
| rs437582745 | 79 | Q>P | No | EVA | |
| rs454788102 | 93 | I>L | No | EVA | |
| rs207642653 | 98 | V>A | No | EVA | |
| rs433014239 | 107 | L>P | No | EVA | |
| rs449794178 | 108 | N>D | No | EVA | |
| rs469956772 | 108 | N>K | No | EVA | |
| rs435368147 | 109 | N>H | No | EVA | |
| rs455062404 | 109 | N>I | No | EVA | |
| rs475086784 | 113 | S>C | No | EVA | |
| rs454089088 | 114 | I>F | No | EVA | |
| rs471312698 | 114 | I>T | No | EVA | |
| rs445755073 | 136 | V>G | No | EVA | |
| rs439886638 | 154 | Y>H | No | EVA | |
| rs470124935 | 157 | N>D | No | EVA | |
| rs470124935 | 157 | N>Y | No | EVA | |
| rs435565817 | 158 | Q>E | No | EVA | |
| rs456011763 | 159 | L>* | No | EVA | |
| rs466278709 | 168 | L>R | No | EVA | |
| rs434984874 | 171 | E>D | No | EVA | |
| rs451757177 | 173 | D>Y | No | EVA | |
| rs471332187 | 179 | V>D | No | EVA | |
| rs473844485 | 191 | V>D | No | EVA | |
| rs456929842 | 191 | V>F | No | EVA | |
| rs449918843 | 211 | W>R | No | EVA | |
| rs797454723 | 260 | A>V | No | EVA | |
| rs443756689 | 297 | E>G | No | EVA | |
| rs463683085 | 298 | D>G | No | EVA | |
| rs442761776 | 312 | M>I | No | EVA | |
| rs459891991 | 313 | R>I | No | EVA | |
| rs480120655 | 347 | N>T | No | EVA | |
| rs445412983 | 350 | D>E | No | EVA | |
| rs471308599 | 350 | D>Y | No | EVA |
No associated diseases with P04696
No regional properties for P04696
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for P04696 | |||
Functions
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| heterotrimeric G-protein complex | Any of a family of heterotrimeric GTP-binding and hydrolyzing proteins; they belong to a superfamily of GTPases that includes monomeric proteins such as EF-Tu and RAS. Heterotrimeric G-proteins consist of three subunits; the alpha subunit contains the guanine nucleotide binding site and possesses GTPase activity; the beta and gamma subunits are tightly associated and function as a beta-gamma heterodimer; extrinsic plasma membrane proteins (cytoplasmic face) that function as a complex to transduce signals from G protein-coupled receptors to an effector protein. |
| photoreceptor inner segment | The inner segment of a vertebrate photoreceptor containing mitochondria, ribosomes and membranes where opsin molecules are assembled and passed to be part of the outer segment discs. |
| photoreceptor outer segment | The outer segment of a vertebrate photoreceptor that contains a stack of membrane discs embedded with photoreceptor proteins. |
| photoreceptor outer segment membrane | The membrane surrounding the outer segment of a vertebrate photoreceptor. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
| metal ion binding | Binding to a metal ion. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation or inhibition of adenylyl cyclase activity and a subsequent change in the intracellular concentration of cyclic AMP (cAMP). |
| detection of chemical stimulus involved in sensory perception of bitter taste | The series of events required for a bitter taste stimulus to be received and converted to a molecular signal. |
| visual perception | The series of events required for an organism to receive a visual stimulus, convert it to a molecular signal, and recognize and characterize the signal. Visual stimuli are detected in the form of photons and are processed to form an image. |
58 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P04695 | GNAT1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P08239 | GNAO1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P0C7Q4 | GNAT3 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P63097 | GNAI1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P38408 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Bos taurus (Bovine) | PR |
| P04896 | GNAS | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Bos taurus (Bovine) | PR |
| P50147 | GNAI2 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P50146 | GNAI1 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P20353 | Galphai | G protein alpha i subunit | Drosophila melanogaster (Fruit fly) | SS |
| P16378 | Galphao | G protein alpha o subunit | Drosophila melanogaster (Fruit fly) | SS |
| P25157 | cta | Guanine nucleotide-binding protein subunit alpha homolog | Drosophila melanogaster (Fruit fly) | SS |
| P11488 | GNAT1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P09471 | GNAO1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q03113 | GNA12 | Guanine nucleotide-binding protein subunit alpha-12 | Homo sapiens (Human) | SS |
| P04899 | GNAI2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P63096 | GNAI1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | EV |
| P08754 | GNAI3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| A8MTJ3 | GNAT3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q14344 | GNA13 | Guanine nucleotide-binding protein subunit alpha-13 | Homo sapiens (Human) | SS |
| P19086 | GNAZ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19087 | GNAT2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q9DC51 | Gnai3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P27600 | Gna12 | Guanine nucleotide-binding protein subunit alpha-12 | Mus musculus (Mouse) | SS |
| P20612 | Gnat1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P27601 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Mus musculus (Mouse) | SS |
| Q3V3I2 | Gnat3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| O70443 | Gnaz | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P08752 | Gnai2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| B2RSH2 | Gnai1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P18872 | Gnao1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P50149 | Gnat2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P93564 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum tuberosum (Potato) | SS |
| Q63210 | Gna12 | Guanine nucleotide-binding protein subunit alpha-12 | Rattus norvegicus (Rat) | SS |
| P19627 | Gnaz | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P29348 | Gnat3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P04897 | Gnai2 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P59215 | Gnao1 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P08753 | Gnai3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| Q6Q7Y5 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Rattus norvegicus (Rat) | SS |
| Q0DJ33 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Oryza sativa subsp. japonica (Rice) | SS |
| P51875 | goa-1 | Guanine nucleotide-binding protein G | Caenorhabditis elegans | SS |
| Q18434 | odr-3 | Guanine nucleotide-binding protein alpha-17 subunit | Caenorhabditis elegans | SS |
| Q9BIG4 | gpa-10 | Guanine nucleotide-binding protein alpha-10 subunit | Caenorhabditis elegans | PR |
| P28052 | gpa-3 | Guanine nucleotide-binding protein alpha-3 subunit | Caenorhabditis elegans | SS |
| Q9XTB2 | gpa-13 | Guanine nucleotide-binding protein alpha-13 subunit | Caenorhabditis elegans | SS |
| Q9BIG5 | gpa-4 | Guanine nucleotide-binding protein alpha-4 subunit | Caenorhabditis elegans | SS |
| P28051 | gpa-1 | Guanine nucleotide-binding protein alpha-1 subunit | Caenorhabditis elegans | SS |
| Q9N2V6 | gpa-16 | Guanine nucleotide-binding protein alpha-16 subunit | Caenorhabditis elegans | SS |
| P91907 | gpa-15 | Guanine nucleotide-binding protein alpha-15 subunit | Caenorhabditis elegans | SS |
| Q20907 | gpa-8 | Guanine nucleotide-binding protein alpha-8 subunit | Caenorhabditis elegans | PR |
| Q21917 | gpa-7 | Guanine nucleotide-binding protein alpha-7 subunit | Caenorhabditis elegans | SS |
| Q93743 | gpa-6 | Guanine nucleotide-binding protein alpha-6 subunit | Caenorhabditis elegans | SS |
| O76584 | gpa-11 | Guanine nucleotide-binding protein alpha-11 subunit | Caenorhabditis elegans | SS |
| P49084 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| P93163 | GPA2 | Guanine nucleotide-binding protein alpha-2 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| O80462 | XLG1 | Extra-large guanine nucleotide-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P18064 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P26981 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGSGASAEDK | ELAKRSKELE | KKLQEDADKE | AKTVKLLLLG | AGESGKSTIV | KQMKIIHQDG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YSPEECLEYK | AIIYGNVLQS | ILAIIRAMPT | LGIDYAEVSC | VDNGRQLNNL | ADSIEEGTMP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PELVEVIRKL | WKDGGVQACF | DRAAEYQLND | SASYYLNQLD | RITAPDYLPN | EQDVLRSRVK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TTGIIETKFS | VKDLNFRMFD | VGGQRSERKK | WIHCFEGVTC | IIFCAALSAY | DMVLVEDDEV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NRMHESLHLF | NSICNHKFFA | ATSIVLFLNK | KDLFEEKIKK | VHLSICFPEY | DGNNSYEDAG |
| 310 | 320 | 330 | 340 | 350 | |
| NYIKSQFLDL | NMRKDVKEIY | SHMTCATDTQ | NVKFVFDAVT | DIIIKENLKD | CGLF |