P00523
Gene name |
SRC |
Protein name |
Proto-oncogene tyrosine-protein kinase Src |
Names |
Proto-oncogene c-Src, pp60c-src, p60-Src |
Species |
Gallus gallus (Chicken) |
KEGG Pathway |
gga:396442 |
EC number |
2.7.10.2: Protein-tyrosine kinases |
Protein Class |
TYROSINE-PROTEIN KINASE (PTHR24418) |
Descriptions
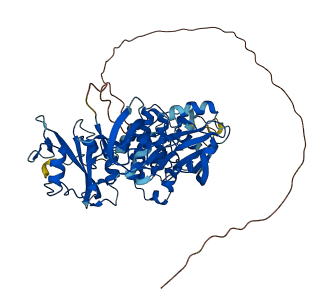
Src is a non-receptor protein tyrosine kinase involved in a variety of signal transduction pathways. SH3 and SH2 domains are assembled on the backside of the kinases and SH2 domain binds to phosphorylated Y527 in C-terminal regulatory tail and the SH3 domain binds to the linker (proline-motif) connecting the SH2 and kinase domains. The binding of pY527 to SH2 domain clamps the SH3-SH2 tandem in the autoinhibitory conformation. Disruption of either SH2-pY527 or SH3-linker interaction disassembles the autoinhibitory conformations and eventually leads to the activation of Src kinases.
Autoinhibitory domains (AIDs)
Target domain |
267-520 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
Structural analysis |
Target domain |
267-520 (Protein kinase domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
Structural analysis |
Target domain |
267-520 (Protein kinase domain) |
Relief mechanism |
|
Assay |
Structural analysis |
Target domain |
267-520 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
Structural analysis |
Accessory elements
403-427 (Activation loop from InterPro)
Target domain |
267-520 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Williams JC et al. (1997) "The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions", Journal of molecular biology, 274, 757-75
- Boggon TJ et al. (2004) "Structure and regulation of Src family kinases", Oncogene, 23, 7918-27
- Meng Y et al. (2014) "Locking the active conformation of c-Src kinase through the phosphorylation of the activation loop", Journal of molecular biology, 426, 423-35
- Register AC et al. (2014) "SH2-catalytic domain linker heterogeneity influences allosteric coupling across the SFK family", Biochemistry, 53, 6910-23
Autoinhibited structure
Activated structure
138 structures for P00523
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1F1W | X-ray | 210 A | A | 145-247 | PDB |
| 1F2F | X-ray | 170 A | A | 145-247 | PDB |
| 1NLO | NMR | - | C | 81-140 | PDB |
| 1NLP | NMR | - | C | 81-140 | PDB |
| 1P13 | X-ray | 163 A | A/B | 145-246 | PDB |
| 1PRL | NMR | - | C | 77-140 | PDB |
| 1PRM | NMR | - | C | 77-140 | PDB |
| 1RLP | NMR | - | C | 77-140 | PDB |
| 1RLQ | NMR | - | C | 77-140 | PDB |
| 1SRL | NMR | - | A | 77-140 | PDB |
| 1SRM | NMR | - | A | 77-140 | PDB |
| 2HWO | X-ray | 250 A | A/B | 251-533 | PDB |
| 2HWP | X-ray | 248 A | A/B | 251-533 | PDB |
| 2OIQ | X-ray | 207 A | A/B | 251-533 | PDB |
| 2PTK | X-ray | 235 A | A | 81-533 | PDB |
| 2QI8 | X-ray | 232 A | A/B | 251-533 | PDB |
| 2QLQ | X-ray | 233 A | A/B | 251-533 | PDB |
| 2QQ7 | X-ray | 238 A | A/B | 251-533 | PDB |
| 3D7T | X-ray | 290 A | B | 251-533 | PDB |
| 3D7U | X-ray | 411 A | B/D | 260-523 | PDB |
| 3DQW | X-ray | 202 A | A/B/C/D | 251-533 | PDB |
| 3DQX | X-ray | 230 A | A/B | 251-533 | PDB |
| 3EL7 | X-ray | 280 A | A | 251-533 | PDB |
| 3EL8 | X-ray | 230 A | A/B | 251-533 | PDB |
| 3EN4 | X-ray | 255 A | A/B | 251-533 | PDB |
| 3EN5 | X-ray | 266 A | A/B | 251-533 | PDB |
| 3EN6 | X-ray | 239 A | A/B | 251-533 | PDB |
| 3EN7 | X-ray | 281 A | A/B | 251-533 | PDB |
| 3F3T | X-ray | 250 A | A/B | 251-533 | PDB |
| 3F3U | X-ray | 250 A | A/B | 251-533 | PDB |
| 3F3V | X-ray | 260 A | A/B | 251-533 | PDB |
| 3F3W | X-ray | 260 A | A/B | 251-533 | PDB |
| 3F6X | X-ray | 235 A | A/B/C/D | 251-533 | PDB |
| 3FJ5 | X-ray | 165 A | A/B | 85-140 | PDB |
| 3G5D | X-ray | 220 A | A/B | 251-533 | PDB |
| 3G6G | X-ray | 231 A | A/B | 251-533 | PDB |
| 3G6H | X-ray | 235 A | A/B | 251-533 | PDB |
| 3GEQ | X-ray | 220 A | A/B | 251-533 | PDB |
| 3LOK | X-ray | 248 A | A/B | 251-533 | PDB |
| 3OEZ | X-ray | 240 A | A/B | 251-533 | PDB |
| 3OF0 | X-ray | 270 A | A/B | 251-533 | PDB |
| 3QLF | X-ray | 275 A | A/B | 251-533 | PDB |
| 3QLG | X-ray | 275 A | A/B | 251-533 | PDB |
| 3SVV | X-ray | 220 A | A/B | 251-533 | PDB |
| 3TZ7 | X-ray | 330 A | A/B | 251-533 | PDB |
| 3TZ8 | X-ray | 270 A | A/B | 251-533 | PDB |
| 3TZ9 | X-ray | 310 A | A/B | 251-533 | PDB |
| 3U4W | X-ray | 190 A | A | 259-533 | PDB |
| 3U51 | X-ray | 224 A | A/B | 259-533 | PDB |
| 3UQF | X-ray | 227 A | A/B | 251-533 | PDB |
| 3UQG | X-ray | 220 A | A/B | 251-533 | PDB |
| 4AGW | X-ray | 260 A | A/B | 251-533 | PDB |
| 4DGG | X-ray | 265 A | A/B | 251-533 | PDB |
| 4FIC | X-ray | 250 A | A/B | 251-533 | PDB |
| 4HVU | X-ray | 098 A | A | 85-141 | PDB |
| 4HVV | X-ray | 110 A | A | 85-140 | PDB |
| 4HVW | X-ray | 098 A | A | 85-141 | PDB |
| 4JZ3 | X-ray | 185 A | A | 85-141 | PDB |
| 4JZ4 | X-ray | 156 A | A/B | 85-141 | PDB |
| 4LE9 | X-ray | 134 A | A | 85-141 | PDB |
| 4LGG | X-ray | 241 A | A/B | 264-533 | PDB |
| 4LGH | X-ray | 284 A | A/B | 257-533 | PDB |
| 4MCV | X-ray | 273 A | A/B | 256-533 | PDB |
| 4O2P | X-ray | 210 A | A/B | 251-533 | PDB |
| 4OML | X-ray | 160 A | A | 85-141 | PDB |
| 4OMM | X-ray | 190 A | A | 85-140 | PDB |
| 4OMN | X-ray | 150 A | A | 85-140 | PDB |
| 4OMO | X-ray | 104 A | A/B | 85-141 | PDB |
| 4OMP | X-ray | 200 A | A | 85-139 | PDB |
| 4OMQ | X-ray | 200 A | A | 85-140 | PDB |
| 4QT7 | X-ray | 155 A | A | 85-141 | PDB |
| 4RTU | X-ray | 245 A | A | 85-141 | PDB |
| 4RTV | X-ray | 137 A | A | 85-141 | PDB |
| 4RTW | X-ray | 124 A | A/C | 85-141 | PDB |
| 4RTX | X-ray | 132 A | A/B/C/D | 85-141 | PDB |
| 4RTY | X-ray | 128 A | A | 85-141 | PDB |
| 4RTZ | X-ray | 098 A | A | 85-141 | PDB |
| 4U5J | X-ray | 226 A | A/B | 251-533 | PDB |
| 4YBJ | X-ray | 261 A | A/B | 251-533 | PDB |
| 4YBK | X-ray | 250 A | A | 251-533 | PDB |
| 5BMM | X-ray | 250 A | A/B | 251-533 | PDB |
| 5D10 | X-ray | 270 A | A/B | 251-533 | PDB |
| 5D11 | X-ray | 230 A | A/B | 251-533 | PDB |
| 5D12 | X-ray | 300 A | A/B | 251-533 | PDB |
| 5EC7 | X-ray | 165 A | A/B/C | 85-140 | PDB |
| 5ECA | X-ray | 116 A | A | 85-141 | PDB |
| 5I11 | X-ray | 195 A | A | 85-141 | PDB |
| 5J5S | X-ray | 215 A | A/B | 251-533 | PDB |
| 5K9I | X-ray | 250 A | A/B | 251-533 | PDB |
| 5OAV | X-ray | 095 A | A/C | 85-141 | PDB |
| 5OB0 | X-ray | 117 A | A | 85-141 | PDB |
| 5OB1 | X-ray | 117 A | A | 85-141 | PDB |
| 5OB2 | X-ray | 180 A | A/C | 85-141 | PDB |
| 5SWH | X-ray | 250 A | A/B | 252-533 | PDB |
| 5SYS | X-ray | 280 A | A/B | 251-533 | PDB |
| 5T0P | X-ray | 250 A | A/B | 251-533 | PDB |
| 5TEH | X-ray | 299 A | A/B | 251-533 | PDB |
| 5XP5 | X-ray | 210 A | A/B | 251-533 | PDB |
| 5XP7 | X-ray | 201 A | A/B | 251-533 | PDB |
| 6HVE | X-ray | 190 A | A/B | 251-533 | PDB |
| 6HVF | X-ray | 210 A | A/B | 251-533 | PDB |
| 6L8L | X-ray | 289 A | A/B/C/D | 251-533 | PDB |
| 6WIW | X-ray | 230 A | A/B | 251-533 | PDB |
| 6XVM | X-ray | 090 A | A/B/C/D | 82-141 | PDB |
| 6XVN | X-ray | 170 A | A/B | 82-141 | PDB |
| 6XVO | X-ray | 170 A | A | 82-141 | PDB |
| 6XX2 | X-ray | 125 A | A | 85-141 | PDB |
| 6XX3 | X-ray | 136 A | A | 85-141 | PDB |
| 6XX4 | X-ray | 105 A | A | 85-141 | PDB |
| 6XX5 | X-ray | 130 A | A | 85-141 | PDB |
| 7A30 | X-ray | 167 A | A | 82-141 | PDB |
| 7A31 | X-ray | 094 A | A/B | 82-141 | PDB |
| 7A32 | X-ray | 115 A | A/B/C/D | 82-141 | PDB |
| 7A33 | X-ray | 096 A | A/B | 82-141 | PDB |
| 7A34 | X-ray | 185 A | A | 82-141 | PDB |
| 7A35 | X-ray | 131 A | A/B | 82-141 | PDB |
| 7A36 | X-ray | 150 A | A/B | 82-141 | PDB |
| 7A37 | X-ray | 152 A | A/B | 82-141 | PDB |
| 7A38 | X-ray | 162 A | A/B | 82-141 | PDB |
| 7A39 | X-ray | 165 A | A/B | 82-141 | PDB |
| 7A3A | X-ray | 180 A | A | 82-141 | PDB |
| 7A3B | X-ray | 191 A | A | 82-141 | PDB |
| 7A3C | X-ray | 180 A | A/B/C/D | 81-141 | PDB |
| 7A3D | X-ray | 220 A | A/B | 82-141 | PDB |
| 7A3E | X-ray | 152 A | A | 81-141 | PDB |
| 7AH3 | X-ray | 195 A | A/B | 251-533 | PDB |
| 7D57 | X-ray | 210 A | A/B | 251-533 | PDB |
| 7D5O | X-ray | 269 A | A/B | 251-533 | PDB |
| 7NER | X-ray | 155 A | A | 81-141 | PDB |
| 7NES | X-ray | 135 A | A | 81-141 | PDB |
| 7NET | X-ray | 150 A | A/B | 81-141 | PDB |
| 7PVW | X-ray | 150 A | A/B | 82-141 | PDB |
| 7PVX | X-ray | 143 A | A/C | 82-141 | PDB |
| 7PVY | X-ray | 140 A | A | 81-141 | PDB |
| 7PVZ | X-ray | 200 A | A/B | 81-141 | PDB |
| 7PW0 | X-ray | 170 A | A/B/C/D/E/F/G/H | 82-141 | PDB |
| 7WF5 | X-ray | 180 A | A/B | 251-533 | PDB |
| AF-P00523-F1 | Predicted | AlphaFoldDB |
25 variants for P00523
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs737050630 | 12 | S>T | No | Ensembl | |
| rs733270406 | 25 | H>P | No | Ensembl | |
| rs733996397 | 26 | H>P | No | Ensembl | |
| rs737461578 | 29 | F>S | No | Ensembl | |
| rs732727338 | 32 | S>P | No | Ensembl | |
| rs741327316 | 44 | H>P | No | Ensembl | |
| rs732500703 | 46 | T>P | No | Ensembl | |
| rs740695096 | 57 | E>A | No | Ensembl | |
| rs731563742 | 66 | T>P | No | Ensembl | |
| rs1059459076 | 72 | S>P | No | Ensembl | |
| rs735146349 | 148 | W>G | No | Ensembl | |
| rs737532680 | 158 | S>P | No | Ensembl | |
| rs732826274 | 176 | E>G | No | Ensembl | |
| rs738256696 | 215 | T>P | No | Ensembl | |
| rs740360366 | 216 | S>P | No | Ensembl | |
| rs734104709 | 239 | H>P | No | Ensembl | |
| rs740661507 | 247 | T>P | No | Ensembl | |
| rs733200424 | 248 | S>P | No | Ensembl | |
| rs733873798 | 255 | L>P | No | Ensembl | |
| rs741442204 | 270 | E>G | No | Ensembl | |
| rs735445953 | 369 | Q>R | No | Ensembl | |
| rs731409743 | 394 | V>G | No | Ensembl | |
| rs733677250 | 498 | C>G | No | Ensembl | |
| rs741234755 | 501 | K>E | No | Ensembl | |
| rs730986301 | 521 | T>P | No | Ensembl |
No associated diseases with P00523
7 regional properties for P00523
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 267 - 520 | IPR000719 |
| domain | SH2 domain | 146 - 245 | IPR000980 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 268 - 515 | IPR001245 |
| domain | SH3 domain | 81 - 142 | IPR001452 |
| active_site | Tyrosine-protein kinase, active site | 382 - 394 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 273 - 295 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 267 - 516 | IPR020635 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.2 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24418 | TYROSINE-PROTEIN KINASE |
| PANTHER Subfamily | PTHR24418:SF53 | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC |
| PANTHER Protein Class | non-receptor tyrosine protein kinase | |
| PANTHER Pathway Category |
Gonadotropin-releasing hormone receptor pathway SRC Angiogenesis Src CCKR signaling map p62 SRC Integrin signalling pathway Src CCKR signaling map SRC @Galphaq Cadherin signaling pathway Src CCKR signaling map SRC CCKR signaling map p60 SRC Parkinson disease Src kinase CCKR signaling map p54 SRC |
|
12 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell junction | A cellular component that forms a specialized region of connection between two or more cells, or between a cell and the extracellular matrix, or between two membrane-bound components of a cell, such as flagella. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| extrinsic component of cytoplasmic side of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to its cytoplasmic surface, but not integrated into the hydrophobic region. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| mitochondrial inner membrane | The inner, i.e. lumen-facing, lipid bilayer of the mitochondrial envelope. It is highly folded to form cristae. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| connexin binding | Binding to a connexin, any of a group of related proteins that assemble to form gap junctions. |
| growth factor receptor binding | Binding to a growth factor receptor. |
| heme binding | Binding to a heme, a compound composed of iron complexed in a porphyrin (tetrapyrrole) ring. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction: ATP + protein L-tyrosine = ADP + protein L-tyrosine phosphate by a non-membrane spanning protein. |
| protein phosphatase binding | Binding to a protein phosphatase. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| bone resorption | The process in which specialized cells known as osteoclasts degrade the organic and inorganic portions of bone, and endocytose and transport the degradation products. |
| cell adhesion | The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules. |
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| epidermal growth factor receptor signaling pathway | The series of molecular signals initiated by binding of a ligand to the tyrosine kinase receptor EGFR (ERBB1) on the surface of a cell. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| negative regulation of extrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of extrinsic apoptotic signaling pathway. |
| negative regulation of intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of intrinsic apoptotic signaling pathway. |
| osteoclast development | The process whose specific outcome is the progression of a osteoclast from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate. An osteoclast is a specialized phagocytic cell associated with the absorption and removal of the mineralized matrix of bone tissue. |
| progesterone receptor signaling pathway | The series of molecular signals initiated by progesterone binding to its receptor in the cytoplasm. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
81 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A0JNB0 | FYN | Tyrosine-protein kinase Fyn | Bos taurus (Bovine) | SS |
| Q0VBZ0 | CSK | Tyrosine-protein kinase CSK | Bos taurus (Bovine) | SS |
| Q3ZC95 | BTK | Tyrosine-protein kinase | Bos taurus (Bovine) | EV SS |
| Q75R65 | JAK2 | Tyrosine-protein kinase JAK2 | Gallus gallus (Chicken) | SS |
| F1N9Y5 | SYK | Tyrosine-protein kinase SYK | Gallus gallus (Chicken) | SS |
| P09324 | YES1 | Tyrosine-protein kinase Yes | Gallus gallus (Chicken) | SS |
| P41239 | CSK | Tyrosine-protein kinase CSK | Gallus gallus (Chicken) | SS |
| Q02977 | YRK | Proto-oncogene tyrosine-protein kinase Yrk | Gallus gallus (Chicken) | SS |
| Q00944 | PTK2 | Focal adhesion kinase 1 | Gallus gallus (Chicken) | EV |
| P42683 | LCK | Proto-oncogene tyrosine-protein kinase LCK | Gallus gallus (Chicken) | SS |
| Q05876 | FYN | Tyrosine-protein kinase Fyn | Gallus gallus (Chicken) | SS |
| Q8JH64 | BTK | Tyrosine-protein kinase BTK | Gallus gallus (Chicken) | SS |
| Q24592 | hop | Tyrosine-protein kinase hopscotch | Drosophila melanogaster (Fruit fly) | PR |
| P08630 | Btk | Tyrosine-protein kinase Btk | Drosophila melanogaster (Fruit fly) | SS |
| P00528 | Src64B | Tyrosine-protein kinase Src64B | Drosophila melanogaster (Fruit fly) | SS |
| Q9V9J3 | Src42A | Tyrosine-protein kinase Src42A | Drosophila melanogaster (Fruit fly) | SS |
| P41240 | CSK | Tyrosine-protein kinase CSK | Homo sapiens (Human) | SS |
| P51451 | BLK | Tyrosine-protein kinase Blk | Homo sapiens (Human) | SS |
| P06239 | LCK | Tyrosine-protein kinase Lck | Homo sapiens (Human) | EV |
| P23458 | JAK1 | Tyrosine-protein kinase JAK1 | Homo sapiens (Human) | SS |
| P06241 | FYN | Tyrosine-protein kinase Fyn | Homo sapiens (Human) | SS |
| P51813 | BMX | Cytoplasmic tyrosine-protein kinase BMX | Homo sapiens (Human) | SS |
| P09769 | FGR | Tyrosine-protein kinase Fgr | Homo sapiens (Human) | SS |
| P42680 | TEC | Tyrosine-protein kinase Tec | Homo sapiens (Human) | SS |
| O60674 | JAK2 | Tyrosine-protein kinase JAK2 | Homo sapiens (Human) | EV |
| P42679 | MATK | Megakaryocyte-associated tyrosine-protein kinase | Homo sapiens (Human) | SS |
| P52333 | JAK3 | Tyrosine-protein kinase JAK3 | Homo sapiens (Human) | SS |
| Q08881 | ITK | Tyrosine-protein kinase ITK/TSK | Homo sapiens (Human) | EV |
| P07948 | LYN | Tyrosine-protein kinase Lyn | Homo sapiens (Human) | SS |
| P29597 | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | Homo sapiens (Human) | EV |
| Q13882 | PTK6 | Protein-tyrosine kinase 6 | Homo sapiens (Human) | EV |
| P08631 | HCK | Tyrosine-protein kinase HCK | Homo sapiens (Human) | EV |
| P07947 | YES1 | Tyrosine-protein kinase Yes | Homo sapiens (Human) | SS |
| P42685 | FRK | Tyrosine-protein kinase FRK | Homo sapiens (Human) | EV |
| Q06187 | BTK | Tyrosine-protein kinase BTK | Homo sapiens (Human) | EV |
| P12931 | SRC | Proto-oncogene tyrosine-protein kinase Src | Homo sapiens (Human) | EV |
| Q9R117 | Tyk2 | Non-receptor tyrosine-protein kinase TYK2 | Mus musculus (Mouse) | SS |
| P08103 | Hck | Tyrosine-protein kinase HCK | Mus musculus (Mouse) | SS |
| P16277 | Blk | Tyrosine-protein kinase Blk | Mus musculus (Mouse) | SS |
| Q62270 | Srms | Tyrosine-protein kinase Srms | Mus musculus (Mouse) | SS |
| Q64434 | Ptk6 | Protein-tyrosine kinase 6 | Mus musculus (Mouse) | SS |
| P14234 | Fgr | Tyrosine-protein kinase Fgr | Mus musculus (Mouse) | SS |
| P35991 | Btk | Tyrosine-protein kinase BTK | Mus musculus (Mouse) | EV |
| P41241 | Csk | Tyrosine-protein kinase CSK | Mus musculus (Mouse) | EV |
| P25911 | Lyn | Tyrosine-protein kinase Lyn | Mus musculus (Mouse) | EV |
| Q62137 | Jak3 | Tyrosine-protein kinase JAK3 | Mus musculus (Mouse) | SS |
| Q62120 | Jak2 | Tyrosine-protein kinase JAK2 | Mus musculus (Mouse) | EV |
| P06240 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Mus musculus (Mouse) | SS |
| P24604 | Tec | Tyrosine-protein kinase Tec | Mus musculus (Mouse) | SS |
| Q04736 | Yes1 | Tyrosine-protein kinase Yes | Mus musculus (Mouse) | SS |
| P39688 | Fyn | Tyrosine-protein kinase Fyn | Mus musculus (Mouse) | SS |
| P52332 | Jak1 | Tyrosine-protein kinase JAK1 | Mus musculus (Mouse) | SS |
| Q03526 | Itk | Tyrosine-protein kinase ITK/TSK | Mus musculus (Mouse) | SS |
| P41242 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q922K9 | Frk | Tyrosine-protein kinase FRK | Mus musculus (Mouse) | SS |
| P05480 | Src | Proto-oncogene tyrosine-protein kinase Src | Mus musculus (Mouse) | EV |
| A1Y2K1 | FYN | Tyrosine-protein kinase Fyn | Sus scrofa (Pig) | SS |
| O19064 | JAK2 | Tyrosine-protein kinase JAK2 | Sus scrofa (Pig) | SS |
| Q62662 | Frk | Tyrosine-protein kinase FRK | Rattus norvegicus (Rat) | SS |
| Q62844 | Fyn | Tyrosine-protein kinase Fyn | Rattus norvegicus (Rat) | SS |
| Q07014 | Lyn | Tyrosine-protein kinase Lyn | Rattus norvegicus (Rat) | SS |
| P50545 | Hck | Tyrosine-protein kinase HCK | Rattus norvegicus (Rat) | SS |
| Q01621 | Lck | Proto-oncogene tyrosine-protein kinase LCK | Rattus norvegicus (Rat) | SS |
| Q6P6U0 | Fgr | Tyrosine-protein kinase Fgr | Rattus norvegicus (Rat) | SS |
| Q62689 | Jak2 | Tyrosine-protein kinase JAK2 | Rattus norvegicus (Rat) | SS |
| Q63272 | Jak3 | Tyrosine-protein kinase JAK3 | Rattus norvegicus (Rat) | SS |
| P32577 | Csk | Tyrosine-protein kinase CSK | Rattus norvegicus (Rat) | SS |
| P41243 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Rattus norvegicus (Rat) | SS |
| F1LM93 | Yes1 | Tyrosine-protein kinase Yes | Rattus norvegicus (Rat) | SS |
| Q9WUD9 | Src | Proto-oncogene tyrosine-protein kinase Src | Rattus norvegicus (Rat) | SS |
| G5ECJ6 | csk-1 | Tyrosine-protein kinase csk-1 | Caenorhabditis elegans | SS |
| O45539 | src-2 | Tyrosine protein-kinase src-2 | Caenorhabditis elegans | SS |
| G5EE56 | src-1 | Tyrosine protein-kinase src-1 | Caenorhabditis elegans | SS |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| A1A5H8 | yes1 | Tyrosine-protein kinase yes | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| F1RDG9 | fynb | Tyrosine-protein kinase fynb | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O12990 | jak1 | Tyrosine-protein kinase JAK1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q6EWH2 | fyna | Tyrosine-protein kinase fyna | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q1JPZ3 | src | Proto-oncogene tyrosine-protein kinase Src | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGSSKSKPKD | PSQRRRSLEP | PDSTHHGGFP | ASQTPNKTAA | PDTHRTPSRS | FGTVATEPKL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FGGFNTSDTV | TSPQRAGALA | GGVTTFVALY | DYESRTETDL | SFKKGERLQI | VNNTEGDWWL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AHSLTTGQTG | YIPSNYVAPS | DSIQAEEWYF | GKITRRESER | LLLNPENPRG | TFLVRESETT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KGAYCLSVSD | FDNAKGLNVK | HYKIRKLDSG | GFYITSRTQF | SSLQQLVAYY | SKHADGLCHR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LTNVCPTSKP | QTQGLAKDAW | EIPRESLRLE | VKLGQGCFGE | VWMGTWNGTT | RVAIKTLKPG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TMSPEAFLQE | AQVMKKLRHE | KLVQLYAVVS | EEPIYIVTEY | MSKGSLLDFL | KGEMGKYLRL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PQLVDMAAQI | ASGMAYVERM | NYVHRDLRAA | NILVGENLVC | KVADFGLARL | IEDNEYTARQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GAKFPIKWTA | PEAALYGRFT | IKSDVWSFGI | LLTELTTKGR | VPYPGMVNRE | VLDQVERGYR |
| 490 | 500 | 510 | 520 | 530 | |
| MPCPPECPES | LHDLMCQCWR | KDPEERPTFE | YLQAFLEDYF | TSTEPQYQPG | ENL |