P00514
Gene name |
PRKAR1A |
Protein name |
cAMP-dependent protein kinase type I-alpha regulatory subunit |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:615074 |
EC number |
|
Protein Class |
CAMP-DEPENDENT PROTEIN KINASE REGULATORY CHAIN (PTHR11635) |
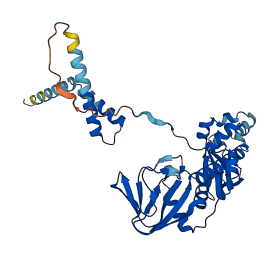
Descriptions
PKA is a major intracellular receptor of cAMP and modulates diverse cellular signalings. The autoinhibitory sequence of the regulatory subunit may interact with the catalytic kinase domain and keeping the catalytic subunit inactive by blocking the active site in the absence of cAMP.
Autoinhibitory domains (AIDs)
Target domain |
136-252 (Cyclic nucleotide-binding domain) |
Relief mechanism |
Ligand binding |
Assay |
Mutagenesis experiment |
Accessory elements
No accessory elements
References
- Turnham RE et al. (2016) "Protein kinase A catalytic subunit isoform PRKACA; History, function and physiology", Gene, 577, 101-8
- Scott JD (1991) "Cyclic nucleotide-dependent protein kinases", Pharmacology & therapeutics, 50, 123-45
- Diskar M et al. (2007) "Molecular basis for isoform-specific autoregulation of protein kinase A", Cellular signalling, 19, 2024-34
Autoinhibited structure

Activated structure

23 structures for P00514
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1NE4 | X-ray | 240 A | A | 95-377 | PDB |
| 1NE6 | X-ray | 230 A | A | 95-377 | PDB |
| 1RGS | X-ray | 280 A | A | 93-380 | PDB |
| 1RL3 | X-ray | 270 A | A/B | 93-380 | PDB |
| 2EZW | NMR | - | A/B | 13-62 | PDB |
| 2QCS | X-ray | 220 A | B | 91-380 | PDB |
| 3FHI | X-ray | 200 A | B | 92-245 | PDB |
| 3IIA | X-ray | 270 A | A | 92-245 | PDB |
| 3IM3 | X-ray | 200 A | A | 13-62 | PDB |
| 3IM4 | X-ray | 228 A | A/B | 13-62 | PDB |
| 3PLQ | X-ray | 230 A | A | 92-245 | PDB |
| 3PNA | X-ray | 150 A | A/B | 92-245 | PDB |
| 3PVB | X-ray | 330 A | B | 85-244 | PDB |
| 4JV4 | X-ray | 295 A | A | 93-380 | PDB |
| 4MX3 | X-ray | 388 A | A/B | 2-380 | PDB |
| 4X6R | X-ray | 240 A | B | 91-380 | PDB |
| 5HVZ | X-ray | 200 A | A/B | 13-62 | PDB |
| 5JR7 | X-ray | 356 A | B/D | 92-366 | PDB |
| 6BYR | X-ray | 366 A | B/D | 2-380 | PDB |
| 6BYS | X-ray | 475 A | B/D/F/H | 2-380 | PDB |
| 6NO7 | X-ray | 355 A | B/D/F/H | 1-380 | PDB |
| 7LZ4 | X-ray | 416 A | A/B/C/D/E/F/G/H | 109-377 | PDB |
| AF-P00514-F1 | Predicted | AlphaFoldDB |
106 variants for P00514
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs135283999 | 6 | T>P | No | Ensembl | |
| rs481459873 | 7 | A>D | No | Ensembl | |
| rs447778005 | 12 | R>C | No | Ensembl | |
| rs477450926 | 14 | L>I | No | Ensembl | |
| rs477450926 | 14 | L>V | No | Ensembl | |
| rs483328506 | 16 | E>D | No | Ensembl | |
| rs442441595 | 25 | N>T | No | Ensembl | |
| rs472166095 | 28 | A>E | No | Ensembl | |
| rs453840374 | 33 | S>P | No | Ensembl | |
| rs438466416 | 39 | T>P | No | Ensembl | |
| rs471270757 | 47 | A>V | No | Ensembl | |
| rs456195014 | 48 | F>C | No | Ensembl | |
| rs467724166 | 58 | K>E | No | Ensembl | |
| rs440471181 | 60 | E>K | No | Ensembl | |
| rs473269570 | 61 | A>E | No | Ensembl | |
| rs451812312 | 64 | I>L | No | Ensembl | |
| rs433325930 | 64 | I>T | No | Ensembl | |
| rs469698250 | 65 | Q>K | No | Ensembl | |
| rs457630857 | 66 | N>H | No | Ensembl | |
| rs435225321 | 66 | N>T | No | Ensembl | |
| rs468196738 | 70 | A>E | No | Ensembl | |
| rs1114625527 | 71 | G>S | No | Ensembl | |
| rs446418554 | 72 | S>R | No | Ensembl | |
| rs1118113689 | 73 | R>C | No | Ensembl | |
| rs464229917 | 74 | A>P | No | Ensembl | |
| rs463663326 | 75 | D>A | No | Ensembl | |
| rs448369233 | 75 | D>E | No | Ensembl | |
| rs482002750 | 75 | D>Y | No | Ensembl | |
| rs481331472 | 77 | R>G | No | Ensembl | |
| rs440334132 | 78 | E>V | No | Ensembl | |
| rs473331171 | 81 | I>L | No | Ensembl | |
| rs458062676 | 81 | I>T | No | Ensembl | |
| rs439802417 | 83 | P>Q | No | Ensembl | |
| rs457421949 | 84 | P>T | No | Ensembl | |
| rs475274693 | 85 | P>H | No | Ensembl | |
| rs435898485 | 85 | P>T | No | Ensembl | |
| rs452951649 | 87 | N>T | No | Ensembl | |
| rs464119704 | 89 | V>A | No | Ensembl | |
| rs464119704 | 89 | V>G | No | Ensembl | |
| rs445669589 | 90 | V>A | No | Ensembl | |
| rs445669589 | 90 | V>G | No | Ensembl | |
| rs437029318 | 91 | K>E | No | Ensembl | |
| rs470091793 | 91 | K>T | No | Ensembl | |
| rs481242171 | 92 | G>V | No | Ensembl | |
| rs446865962 | 93 | R>G | No | Ensembl | |
| rs439559484 | 96 | R>H | No | Ensembl | |
| rs464047029 | 97 | G>R | No | Ensembl | |
| rs475206735 | 99 | I>L | No | Ensembl | |
| rs434423581 | 99 | I>M | No | Ensembl | |
| rs453542528 | 99 | I>S | No | Ensembl | |
| rs475206735 | 99 | I>V | No | Ensembl | |
| rs470683348 | 101 | A>G | No | Ensembl | |
| rs436775944 | 102 | E>* | No | Ensembl | |
| rs469796359 | 102 | E>G | No | Ensembl | |
| rs454792281 | 103 | V>G | No | Ensembl | |
| rs436303026 | 105 | T>P | No | Ensembl | |
| rs465760939 | 106 | E>A | No | Ensembl | |
| rs465760939 | 106 | E>G | No | Ensembl | |
| rs464483571 | 108 | D>E | No | Ensembl | |
| rs447402515 | 108 | D>N | No | Ensembl | |
| rs477055674 | 108 | D>V | No | Ensembl | |
| rs446134893 | 109 | A>P | No | Ensembl | |
| rs446134893 | 109 | A>T | No | Ensembl | |
| rs463739050 | 111 | S>A | No | Ensembl | |
| rs463739050 | 111 | S>T | No | Ensembl | |
| rs442270023 | 112 | Y>D | No | Ensembl | |
| rs481782573 | 112 | Y>F | No | Ensembl | |
| rs459887508 | 114 | R>G | No | Ensembl | |
| rs441434027 | 115 | K>Q | No | Ensembl | |
| rs382810255 | 152 | M>T | No | Ensembl | |
| rs450398153 | 158 | I>M | No | Ensembl | |
| rs440356673 | 230 | Y>* | No | Ensembl | |
| rs466136680 | 247 | E>K | No | Ensembl | |
| rs434855385 | 259 | D>N | No | Ensembl | |
| rs478148752 | 286 | E>K | No | Ensembl | |
| rs462854177 | 286 | E>V | No | Ensembl | |
| rs455003345 | 292 | F>L | No | Ensembl | |
| rs442964601 | 294 | I>F | No | Ensembl | |
| rs472539829 | 294 | I>N | No | Ensembl | |
| rs454106150 | 296 | E>G | No | Ensembl | |
| rs464338138 | 311 | F>V | No | Ensembl | |
| rs450926995 | 326 | I>N | No | Ensembl | |
| rs480388143 | 327 | A>V | No | Ensembl | |
| rs461990638 | 328 | L>M | No | Ensembl | |
| rs440643985 | 332 | R>G | No | Ensembl | |
| rs440643985 | 332 | R>W | No | Ensembl | |
| rs439776530 | 333 | P>H | No | Ensembl | |
| rs458491505 | 333 | P>S | No | Ensembl | |
| rs476033042 | 336 | A>V | No | Ensembl | |
| rs463419223 | 337 | T>P | No | Ensembl | |
| rs441754204 | 340 | A>V | No | Ensembl | |
| rs470862575 | 347 | V>G | No | Ensembl | |
| rs436936450 | 353 | R>G | No | Ensembl | |
| rs469825552 | 354 | F>I | No | Ensembl | |
| rs435656843 | 359 | G>C | No | Ensembl | |
| rs468746689 | 361 | C>G | No | Ensembl | |
| rs446799399 | 362 | S>P | No | Ensembl | |
| rs479891350 | 363 | D>A | No | Ensembl | |
| rs458356259 | 364 | I>N | No | Ensembl | |
| rs458356259 | 364 | I>T | No | Ensembl | |
| rs446352431 | 365 | L>I | No | Ensembl | |
| rs482380598 | 369 | I>L | No | Ensembl | |
| rs463960825 | 372 | Y>C | No | Ensembl | |
| rs441690812 | 376 | V>G | No | Ensembl | |
| rs474692989 | 380 | V>F | No | Ensembl | |
| rs459489001 | 381 | V>C | No | Ensembl |
No associated diseases with P00514
7 regional properties for P00514
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 268 - 521 | IPR000719 |
| domain | SH2 domain | 147 - 253 | IPR000980 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 269 - 515 | IPR001245 |
| domain | SH3 domain | 82 - 143 | IPR001452 |
| active_site | Tyrosine-protein kinase, active site | 383 - 395 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 274 - 296 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 268 - 517 | IPR020635 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11635 | CAMP-DEPENDENT PROTEIN KINASE REGULATORY CHAIN |
| PANTHER Subfamily | PTHR11635:SF129 | CAMP-DEPENDENT PROTEIN KINASE TYPE I-ALPHA REGULATORY SUBUNIT |
| PANTHER Protein Class | kinase modulator | |
| PANTHER Pathway Category |
Metabotropic glutamate receptor group III pathway PKA Muscarinic acetylcholine receptor 2 and 4 signaling pathway PKA GABA-B receptor II signaling PKA Transcription regulation by bZIP transcription factor PKA Metabotropic glutamate receptor group II pathway PKA Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway PKA-r Hedgehog signaling pathway PKA Endothelin signaling pathway PKA Metabotropic glutamate receptor group I pathway PKA |
|
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| axoneme | The bundle of microtubules and associated proteins that forms the core of cilia (also called flagella) in eukaryotic cells and is responsible for their movements. |
| cAMP-dependent protein kinase complex | An enzyme complex, composed of regulatory and catalytic subunits, that catalyzes protein phosphorylation. Inactive forms of the enzyme have two regulatory chains and two catalytic chains; activation by cAMP produces two active catalytic monomers and a regulatory dimer. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| immunological synapse | An area of close contact between a lymphocyte (T-, B-, or natural killer cell) and a target cell formed through the clustering of particular signaling and adhesion molecules and their associated membrane rafts on both the lymphocyte and the target cell and facilitating activation of the lymphocyte, transfer of membrane from the target cell to the lymphocyte, and in some situations killing of the target cell through release of secretory granules and/or death-pathway ligand-receptor interaction. |
| multivesicular body | A type of endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| neuromuscular junction | The junction between the axon of a motor neuron and a muscle fiber. In response to the arrival of action potentials, the presynaptic button releases molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane of the muscle fiber, leading to a change in post-synaptic potential. |
| nucleotide-activated protein kinase complex | A protein complex that possesses nucleotide-dependent protein kinase activity. The nucleotide can be AMP (in S. pombe and human) or ADP (in S. cerevisiae). |
| plasma membrane raft | A membrane raft that is part of the plasma membrane. |
| sperm connecting piece | The segment of the sperm flagellum that attaches to the implantation fossa of the nucleus in the sperm head; from the remnant of the centriole at this point, the axoneme extends throughout the length of the flagellum. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| cAMP binding | Binding to cAMP, the nucleotide cyclic AMP (adenosine 3',5'-cyclophosphate). |
| cAMP-dependent protein kinase inhibitor activity | Binds to and stops, prevents or reduces the activity of a cAMP-dependent protein kinase. |
| identical protein binding | Binding to an identical protein or proteins. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase A catalytic subunit binding | Binding to one or both of the catalytic subunits of protein kinase A. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| cardiac muscle cell proliferation | The expansion of a cardiac muscle cell population by cell division. |
| mesoderm formation | The process that gives rise to the mesoderm. This process pertains to the initial formation of the structure from unspecified parts. |
| negative regulation of activated T cell proliferation | Any process that stops, prevents or reduces the rate or extent of activated T cell proliferation. |
| negative regulation of cAMP-dependent protein kinase activity | Any process that stops, prevents or reduces the frequency, rate or extent of cAMP-dependent protein kinase activity. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| sarcomere organization | The myofibril assembly process that results in the organization of muscle actomyosin into sarcomeres. The sarcomere is the repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs. |
22 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P31322 | PRKAR2B | cAMP-dependent protein kinase type II-beta regulatory subunit | Bos taurus (Bovine) | EV |
| P00515 | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit | Bos taurus (Bovine) | EV |
| Q5ZM91 | PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | Gallus gallus (Chicken) | SS |
| P81900 | Pka-R2 | cAMP-dependent protein kinase type II regulatory subunit | Drosophila melanogaster (Fruit fly) | PR |
| P16905 | Pka-R1 | cAMP-dependent protein kinase type I regulatory subunit | Drosophila melanogaster (Fruit fly) | PR |
| Q96M20 | CNBD2 | Cyclic nucleotide-binding domain-containing protein 2 | Homo sapiens (Human) | PR |
| P13861 | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit | Homo sapiens (Human) | PR |
| P31321 | PRKAR1B | cAMP-dependent protein kinase type I-beta regulatory subunit | Homo sapiens (Human) | EV |
| P31323 | PRKAR2B | cAMP-dependent protein kinase type II-beta regulatory subunit | Homo sapiens (Human) | PR |
| P10644 | PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | Homo sapiens (Human) | EV |
| P12367 | Prkar2a | cAMP-dependent protein kinase type II-alpha regulatory subunit | Mus musculus (Mouse) | PR |
| Q9D5U8 | Cnbd2 | Cyclic nucleotide-binding domain-containing protein 2 | Mus musculus (Mouse) | PR |
| P12849 | Prkar1b | cAMP-dependent protein kinase type I-beta regulatory subunit | Mus musculus (Mouse) | PR |
| P31324 | Prkar2b | cAMP-dependent protein kinase type II-beta regulatory subunit | Mus musculus (Mouse) | PR |
| Q9DBC7 | Prkar1a | cAMP-dependent protein kinase type I-alpha regulatory subunit [Cleaved into: cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed] | Mus musculus (Mouse) | SS |
| P05207 | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit | Sus scrofa (Pig) | SS |
| P07802 | PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | Sus scrofa (Pig) | SS |
| P81377 | Prkar1b | cAMP-dependent protein kinase type I-beta regulatory subunit | Rattus norvegicus (Rat) | PR |
| P09456 | Prkar1a | cAMP-dependent protein kinase type I-alpha regulatory subunit | Rattus norvegicus (Rat) | SS |
| P12368 | Prkar2a | cAMP-dependent protein kinase type II-alpha regulatory subunit | Rattus norvegicus (Rat) | PR |
| P12369 | Prkar2b | cAMP-dependent protein kinase type II-beta regulatory subunit | Rattus norvegicus (Rat) | PR |
| P30625 | kin-2 | cAMP-dependent protein kinase regulatory subunit | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASGTTASEE | ERSLRECELY | VQKHNIQALL | KDSIVQLCTA | RPERPMAFLR | EYFEKLEKEE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AKQIQNLQKA | GSRADSREDE | ISPPPPNPVV | KGRRRRGAIS | AEVYTEEDAA | SYVRKVIPKD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| YKTMAALAKA | IEKNVLFSHL | DDNERSDIFD | AMFPVSFIAG | ETVIQQGDEG | DNFYVIDQGE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MDVYVNNEWA | TSVGEGGSFG | ELALIYGTPR | AATVKAKTNV | KLWGIDRDSY | RRILMGSTLR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KRKMYEEFLS | KVSILESLDK | WERLTVADAL | EPVQFEDGQK | IVVQGEPGDE | FFIILEGSAA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VLQRRSENEE | FVEVGRLGPS | DYFGEIALLM | NRPRAATVVA | RGPLKCVKLD | RPRFERVLGP |
| 370 | |||||
| CSDILKRNIQ | QYNSFVSLSV |