O95376
Gene name |
ARIH2 (ARI2, TRIAD1) |
Protein name |
E3 ubiquitin-protein ligase ARIH2 |
Names |
ARI-2, Protein ariadne-2 homolog, RING-type E3 ubiquitin transferase ARIH2, Triad1 protein |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:10425 |
EC number |
2.3.2.31: Aminoacyltransferases |
Protein Class |
RBR FAMILY RING FINGER AND IBR DOMAIN-CONTAINING (PTHR11685) |
Descriptions
E3 ubiquitin-protein ligase ARIH2 catalyzes ubiquitination of target proteins together with ubiquitin-conjugating enzyme E2 UBE2L3 and acts as an atypical E3 ubiquitin-protein ligase by working together with cullin-5-RING ubiquitin ligase complex and initiating ubiquitination of ECS substrates. ARIH2 is autoinhibited by the ariadne domain, which masks the second RING-type zinc finger that contains the active site and inhibits the E3 activity. Inhibition is relieved upon binding to neddylated cullin-RING ubiquitin ligase complexes, which activate the E3 ligase activity.
Autoinhibitory domains (AIDs)
Target domain |
300-340 (RING2 domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
References
- Kelsall IR et al. (2013) "TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes", The EMBO journal, 32, 2848-60
- Kostrhon S et al. (2021) "CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation", Nature chemical biology, 17, 1075-1083
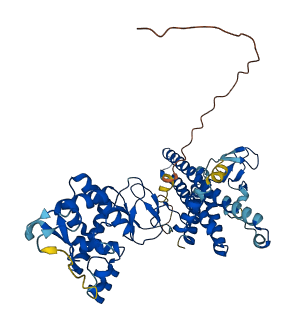
Autoinhibited structure
Activated structure
3 structures for O95376
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 7OD1 | X-ray | 245 A | A/B | 1-493 | PDB |
| 7ONI | EM | 340 A | H | 1-493 | PDB |
| AF-O95376-F1 | Predicted | AlphaFoldDB |
181 variants for O95376
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs146714746 CA2387764 |
5 | M>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA2387765 rs775296209 |
6 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352648771 rs1251780705 |
10 | S>A | No |
ClinGen gnomAD |
|
|
rs1251780705 CA352648749 |
10 | S>P | No |
ClinGen gnomAD |
|
|
CA352648816 rs1338674813 |
11 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs768674285 CA2387767 |
12 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs776299644 CA2387768 |
13 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs200421825 CA73983302 |
16 | D>Y | No |
ClinGen 1000Genomes |
|
|
rs1188970482 CA352649135 |
17 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
CA2387769 rs138105757 |
18 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA352649235 rs1172945432 |
21 | C>R | No |
ClinGen gnomAD |
|
|
rs1553695679 CA2387771 |
21 | C>W | No |
ClinGen Ensembl |
|
|
rs11507 VAR_054105 CA73983339 |
24 | E>K | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs762611632 CA2387777 |
28 | E>D | No |
ClinGen ExAC gnomAD |
|
|
VAR_054106 rs34221642 CA2387778 |
29 | E>D | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA73983389 rs753068638 |
30 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772777170 CA352649684 |
31 | D>H | No |
ClinGen gnomAD |
|
|
rs772777170 CA73983395 |
31 | D>N | No |
ClinGen gnomAD |
|
|
rs150747484 CA2387782 |
32 | P>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2387783 rs754314513 |
32 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA2387784 COSM1045973 rs757397436 |
37 | D>N | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA73983440 rs1021548669 |
40 | V>M | No |
ClinGen Ensembl |
|
|
CA352650167 rs1348383417 |
44 | S>G | No |
ClinGen gnomAD |
|
|
CA2387786 rs746090221 |
44 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746090221 CA352650168 |
44 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1186750262 CA352650251 |
47 | E>G | No |
ClinGen TOPMed |
|
|
rs977408244 CA73983479 |
50 | G>A | No |
ClinGen gnomAD |
|
|
rs779744288 CA2387788 |
50 | G>W | No |
ClinGen ExAC gnomAD |
|
|
CA73983495 rs112345459 |
57 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1191734093 CA352650559 |
57 | E>K | No |
ClinGen gnomAD |
|
|
rs776747405 CA2387791 |
58 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA73983512 rs201971966 |
61 | F>L | No |
ClinGen 1000Genomes |
|
|
CA352650717 rs200385612 |
62 | T>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs200385612 CA2387792 |
62 | T>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2387794 rs772714830 |
66 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1559717167 CA352650831 |
67 | K>Q | No |
ClinGen Ensembl |
|
|
rs762547621 CA2387795 |
67 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA2387796 rs766011857 |
68 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA2387798 rs761068678 |
74 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs1371680328 CA352651129 |
77 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA2387799 rs199731867 |
79 | S>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA352651325 rs1218289551 |
85 | K>N | No |
ClinGen gnomAD |
|
|
rs1285107929 CA352664703 |
88 | H>L | No |
ClinGen gnomAD |
|
|
rs777806461 CA2387836 |
88 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs368954794 CA2387837 |
90 | V>I | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1208090185 CA352664768 |
91 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1044557813 CA74007509 |
94 | I>M | No |
ClinGen Ensembl |
|
|
rs770534282 CA2387838 |
94 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352664997 rs1185909845 |
97 | N>S | No |
ClinGen gnomAD |
|
|
CA352665077 rs1268185468 |
99 | H>Y | No |
ClinGen TOPMed |
|
|
CA352665172 rs1455390747 |
101 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
rs778423872 CA2387859 |
112 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1443845395 CA352666751 |
114 | A>V | No |
ClinGen gnomAD |
|
|
rs757880723 CA2387861 |
116 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA74009687 rs1042764160 |
117 | L>F | No |
ClinGen Ensembl |
|
|
CA74009691 rs904255797 |
117 | L>R | No |
ClinGen Ensembl |
|
|
rs527850935 CA2387862 |
118 | V>F | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs527850935 CA352666847 |
118 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2387863 rs746633869 |
119 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs112979690 CA2387864 |
121 | R>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs773388389 CA2387865 |
121 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA352667017 rs1576443772 |
122 | V>G | No |
ClinGen Ensembl |
|
|
rs749836106 CA2387866 |
124 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs377679410 CA2387867 |
125 | N>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1576443875 CA352667172 |
128 | K>E | No |
ClinGen Ensembl |
|
|
rs1162602317 CA352668603 |
130 | V>D | No |
ClinGen TOPMed |
|
|
rs367621951 CA2387890 |
130 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs761632702 CA2387893 |
133 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2387892 rs761632702 |
133 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs566139192 CA74011450 |
135 | P>H | No |
ClinGen TOPMed gnomAD |
|
|
rs746152958 CA2387895 |
138 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA74011469 rs201979858 |
139 | C>* | No |
ClinGen 1000Genomes |
|
|
CA352669169 rs1292201451 |
144 | Q>H | No |
ClinGen gnomAD |
|
|
rs995636397 CA74011471 |
145 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs554254556 CA2387897 |
147 | R>* | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs554254556 CA2387896 |
147 | R>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1270532962 CA352669230 |
147 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1338452691 CA352669240 |
148 | K>E | No |
ClinGen gnomAD |
|
|
rs1202802481 CA352669285 |
148 | K>N | No |
ClinGen gnomAD |
|
|
CA352669372 rs1220021601 |
150 | N>T | No |
ClinGen TOPMed |
|
|
COSM1753258 rs1033760412 CA74011501 |
153 | S>C | urinary_tract [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA74011502 rs921369653 |
159 | Q>R | No |
ClinGen Ensembl |
|
|
CA74011505 rs932796208 |
162 | R>H | No |
ClinGen gnomAD |
|
|
rs1559827017 CA352669819 |
163 | S>C | No |
ClinGen Ensembl |
|
|
rs1422549007 CA352669831 |
163 | S>N | No |
ClinGen gnomAD |
|
|
CA2387899 rs780747982 |
167 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
rs752393082 CA2387900 |
168 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1296781414 CA352670106 |
169 | C>G | No |
ClinGen TOPMed |
|
|
COSM460782 rs201245336 CA2387902 |
173 | V>I | cervix haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1399500675 CA352670374 |
175 | D>N | No |
ClinGen gnomAD |
|
|
rs1318844980 CA352670441 |
176 | G>S | No |
ClinGen gnomAD |
|
|
CA2387905 rs780202533 |
177 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352670557 rs1305429095 |
178 | G>D | No |
ClinGen gnomAD |
|
|
CA2387907 rs768746772 |
179 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA2387936 rs763886213 |
187 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352672322 rs1336132245 |
187 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA2387935 rs760428061 |
187 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1264418280 CA352672400 |
189 | P>S | No |
ClinGen gnomAD |
|
|
rs753506586 CA2387937 |
191 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA2387938 rs546937892 |
191 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs753506586 CA74012510 |
191 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA2387939 rs766749204 |
192 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs962077158 CA74012522 |
194 | E>D | No |
ClinGen TOPMed |
|
|
CA74012530 rs200216731 |
198 | F>L | No |
ClinGen Ensembl |
|
|
CA352672894 rs1423230644 |
203 | N>D | No |
ClinGen gnomAD |
|
|
rs112266750 CA2387943 |
203 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs529167013 CA74012555 |
211 | R>G | No |
ClinGen 1000Genomes |
|
|
rs778037511 CA352673265 |
211 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2387945 rs778037511 |
211 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352673272 rs1370190130 |
212 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs867071514 CA74012559 |
212 | R>H | No |
ClinGen Ensembl |
|
|
rs1391502628 CA352673405 |
216 | R>K | No |
ClinGen TOPMed |
|
|
rs144088007 CA2387947 |
218 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2387965 rs757464607 |
222 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1486189292 CA352674505 |
237 | I>M | No |
ClinGen gnomAD |
|
|
rs746926650 CA2387970 |
238 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2387969 rs775632661 |
238 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs768290698 CA2387971 |
241 | E>K | No |
ClinGen ExAC TOPMed |
|
|
CA352674699 rs1338501907 |
246 | R>Q | No |
ClinGen Ensembl |
|
|
rs776140482 CA2387972 |
247 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2387973 rs376123571 |
250 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2387975 rs772715889 |
251 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769437830 CA2387974 |
251 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1436550025 CA352674941 |
253 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA352674976 rs767856740 |
254 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA352675010 rs1355027825 |
255 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1172186572 CA352674981 |
255 | V>I | No |
ClinGen TOPMed |
|
|
CA2387994 rs762534832 |
259 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA2387995 rs772431690 |
261 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs980773694 COSM3824095 CA74014661 |
261 | R>H | breast [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs980773694 CA352676637 |
261 | R>L | No |
ClinGen TOPMed |
|
|
rs761197257 CA2387997 |
266 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352677129 rs1472921582 |
274 | R>Q | No |
ClinGen gnomAD |
|
|
rs762017329 CA2388000 |
275 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA2388002 rs750666445 |
278 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs866392304 CA74014688 |
283 | D>N | No |
ClinGen TOPMed |
|
|
CA2388006 rs754796526 |
291 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA352677847 rs1330422321 |
294 | T>A | No |
ClinGen gnomAD |
|
|
rs1228012437 CA352678976 |
300 | C>R | No |
ClinGen gnomAD |
|
|
CA2388024 rs751798787 |
310 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs370396255 CA2388037 |
317 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1252116594 CA352680221 |
320 | H>Y | No |
ClinGen TOPMed |
|
|
rs753773992 CA2388049 |
329 | D>N | No |
ClinGen ExAC gnomAD |
|
|
COSM1183598 CA352683143 rs1188915884 |
342 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA2388050 rs756815478 |
342 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs750091114 CA2388052 |
345 | E>G | No |
ClinGen ExAC gnomAD |
|
|
COSM1172989 CA2388055 rs748510233 |
350 | V>M | oesophagus [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed |
|
rs1287785394 CA352683495 |
353 | S>G | No |
ClinGen TOPMed |
|
|
rs200594843 CA2388058 |
356 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1388853437 CA352683623 |
357 | Q>* | No |
ClinGen TOPMed gnomAD |
|
|
rs1227050558 CA352683687 |
358 | A>V | No |
ClinGen gnomAD |
|
|
rs1576564543 CA352685919 |
376 | N>S | No |
ClinGen Ensembl |
|
|
rs1395797554 CA352686376 |
386 | Y>C | No |
ClinGen gnomAD |
|
|
rs1430538617 CA352686352 |
386 | Y>H | No |
ClinGen TOPMed |
|
|
CA2388072 rs757902535 |
388 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2388074 rs751218573 |
390 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2388076 COSM1045976 rs778218697 |
391 | E>K | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC |
|
CA352686833 rs771517976 |
396 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA352686872 rs1211620874 |
398 | M>T | No |
ClinGen gnomAD |
|
|
CA2388079 rs779542145 |
399 | N>S | No |
ClinGen ExAC |
|
|
rs1237927374 CA352686944 |
400 | N>S | No |
ClinGen gnomAD |
|
|
CA352687100 rs1559867992 |
406 | D>N | No |
ClinGen Ensembl |
|
|
CA74016857 rs76267185 |
409 | Y>D | No |
ClinGen Ensembl |
|
|
rs772190175 CA2388081 |
412 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA74016862 rs918121667 |
415 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs758425518 CA2388103 |
421 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2388107 rs776808107 |
435 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs769571023 CA2388109 |
437 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773081132 CA2388110 |
438 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs111941464 CA74017252 |
440 | K>R | No |
ClinGen Ensembl |
|
|
CA74017559 rs112257171 |
447 | Q>R | No |
ClinGen Ensembl |
|
|
CA352690275 rs373636239 |
460 | K>R | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA74017564 rs373636239 |
460 | K>T | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1559875485 CA352691040 |
478 | I>V | No |
ClinGen Ensembl |
|
|
rs1333235363 CA352691201 |
482 | R>G | No |
ClinGen gnomAD |
|
|
CA2388147 rs749150125 |
482 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA352691310 rs1300024418 |
485 | T>S | No |
ClinGen gnomAD |
|
|
rs1186572296 CA352691590 |
491 | H>Q | No |
ClinGen TOPMed |
|
|
CA2388150 rs745744423 |
492 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771716485 CA2388151 |
493 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs771716485 CA352691640 |
493 | T>N | No |
ClinGen ExAC gnomAD |
No associated diseases with O95376
9 regional properties for O95376
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Zinc finger, RING-type | 139 - 190 | IPR001841-1 |
| domain | Zinc finger, RING-type | 300 - 340 | IPR001841-2 |
| domain | IBR domain | 208 - 270 | IPR002867-1 |
| domain | IBR domain | 278 - 340 | IPR002867-2 |
| conserved_site | Zinc finger, RING-type, conserved site | 318 - 327 | IPR017907 |
| domain | TRIAD supradomain | 135 - 344 | IPR044066 |
| domain | Ariadne domain | 353 - 470 | IPR045840 |
| domain | E3 ubiquitin-protein ligase ARIH2, BRcat domain | 217 - 288 | IPR047555 |
| domain | E3 ubiquitin-protein ligase TRIAD1, Rcat domain | 292 - 347 | IPR047556 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.31 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11685 | RBR FAMILY RING FINGER AND IBR DOMAIN-CONTAINING |
| PANTHER Subfamily | PTHR11685:SF210 | E3 UBIQUITIN-PROTEIN LIGASE ARIH2 |
| PANTHER Protein Class |
ubiquitin-protein ligase
protein modifying enzyme |
|
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| ubiquitin ligase complex | A protein complex that includes a ubiquitin-protein ligase and enables ubiquitin protein ligase activity. The complex also contains other proteins that may confer substrate specificity on the complex. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ubiquitin conjugating enzyme binding | Binding to a ubiquitin conjugating enzyme, any of the E2 proteins. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
| zinc ion binding | Binding to a zinc ion (Zn). |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| developmental cell growth | The growth of a cell, where growth contributes to the progression of the cell over time from one condition to another. |
| hematopoietic stem cell proliferation | The expansion of a hematopoietic stem cell population by cell division. A hematopoietic stem cell is a stem cell from which all cells of the lymphoid and myeloid lineages develop. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| positive regulation of protein targeting to mitochondrion | Any process that activates or increases the frequency, rate or extent of protein targeting to mitochondrion. |
| protein K48-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 48 of the ubiquitin monomers, is added to a protein. K48-linked ubiquitination targets the substrate protein for degradation. |
| protein K63-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 63 of the ubiquitin monomers, is added to a protein. K63-linked ubiquitination does not target the substrate protein for degradation, but is involved in several pathways, notably as a signal to promote error-free DNA postreplication repair. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin group, or multiple ubiquitin groups, to the protein. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VEA3 | ARIH1 | E3 ubiquitin-protein ligase ARIH1 | Bos taurus (Bovine) | SS |
| O76924 | ari-2 | Potential E3 ubiquitin-protein ligase ariadne-2 | Drosophila melanogaster (Fruit fly) | SS |
| Q94981 | ari-1 | E3 ubiquitin-protein ligase ariadne-1 | Drosophila melanogaster (Fruit fly) | SS |
| Q9NWF9 | RNF216 | E3 ubiquitin-protein ligase RNF216 | Homo sapiens (Human) | PR |
| O60260 | PRKN | E3 ubiquitin-protein ligase parkin | Homo sapiens (Human) | EV |
| Q9Y4X5 | ARIH1 | E3 ubiquitin-protein ligase ARIH1 | Homo sapiens (Human) | EV |
| Q9Z1K6 | Arih2 | E3 ubiquitin-protein ligase ARIH2 | Mus musculus (Mouse) | SS |
| Q9Z1K5 | Arih1 | E3 ubiquitin-protein ligase ARIH1 | Mus musculus (Mouse) | SS |
| Q22431 | ari-2 | Potential E3 ubiquitin-protein ligase ariadne-2 | Caenorhabditis elegans | SS |
| O01965 | ari-1.1 | E3 ubiquitin-protein ligase ari-1.1 | Caenorhabditis elegans | SS |
| B1H1E4 | arih1 | E3 ubiquitin-protein ligase arih1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q6PFJ9 | arih1 | E3 ubiquitin-protein ligase arih1 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q6NW85 | arih1l | E3 ubiquitin-protein ligase arih1l | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSVDMNSQGS | DSNEEDYDPN | CEEEEEEEED | DPGDIEDYYV | GVASDVEQQG | ADAFDPEEYQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FTCLTYKESE | GALNEHMTSL | ASVLKVSHSV | AKLILVNFHW | QVSEILDRYK | SNSAQLLVEA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RVQPNPSKHV | PTSHPPHHCA | VCMQFVRKEN | LLSLACQHQF | CRSCWEQHCS | VLVKDGVGVG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VSCMAQDCPL | RTPEDFVFPL | LPNEELREKY | RRYLFRDYVE | SHYQLQLCPG | ADCPMVIRVQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EPRARRVQCN | RCNEVFCFKC | RQMYHAPTDC | ATIRKWLTKC | ADDSETANYI | SAHTKDCPKC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NICIEKNGGC | NHMQCSKCKH | DFCWMCLGDW | KTHGSEYYEC | SRYKENPDIV | NQSQQAQARE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ALKKYLFYFE | RWENHNKSLQ | LEAQTYQRIH | EKIQERVMNN | LGTWIDWQYL | QNAAKLLAKC |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RYTLQYTYPY | AYYMESGPRK | KLFEYQQAQL | EAEIENLSWK | VERADSYDRG | DLENQMHIAE |
| 490 | |||||
| QRRRTLLKDF | HDT |