O95185
Gene name |
UNC5C (UNC5H3) |
Protein name |
Netrin receptor UNC5C |
Names |
Protein unc-5 homolog 3 , Protein unc-5 homolog C |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:8633 |
EC number |
|
Protein Class |
|
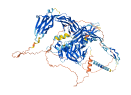
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
528-928 (ZU5-UPA-DD) |
Relief mechanism |
Ligand binding |
Assay |
|
Target domain |
528-928 (ZU5-UPA-DD) |
Relief mechanism |
Ligand binding |
Assay |
|
Target domain |
528-928 (ZU5-UPA-DD) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for O95185
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O95185-F1 | Predicted | AlphaFoldDB |
1034 variants for O95185
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
VAR_081368 rs137875858 |
835 | T>M | AD; increased susceptibility to neuronal cell death [UniProt] | Yes |
UniProt ESP ExAC TOPMed dbSNP gnomAD |
| TCGA novel | 1 | M>? | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757386410 | 3 | K>* | No |
ExAC gnomAD |
|
| rs751983361 | 3 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 4 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764501513 | 4 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1560502608 | 4 | G>D | No | TOPMed | |
| rs892982110 | 6 | R>P | No | TOPMed | |
| rs892982110 | 6 | R>Q | No | TOPMed | |
| rs765835503 | 6 | R>W | No |
ExAC gnomAD |
|
| rs1723151067 | 7 | A>E | No | Ensembl | |
| rs760290190 | 7 | A>P | No |
ExAC gnomAD |
|
| rs771787598 | 8 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1723150751 | 8 | T>I | No | gnomAD | |
| rs372991064 | 9 | A>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs549704340 | 9 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs372991064 | 9 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs745996659 | 11 | R>C | No |
ExAC gnomAD |
|
| rs1365890144 | 11 | R>H | No |
TOPMed gnomAD |
|
| rs1365890144 | 11 | R>L | No |
TOPMed gnomAD |
|
| rs745996659 | 11 | R>S | No |
ExAC gnomAD |
|
| rs1217891652 | 13 | G>A | No |
TOPMed gnomAD |
|
| rs1217891652 | 13 | G>E | No |
TOPMed gnomAD |
|
| rs1313721643 | 14 | L>M | No |
TOPMed gnomAD |
|
| TCGA novel | 15 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs529808046 | 15 | G>R | No |
1000Genomes ExAC gnomAD |
|
| rs757520579 | 17 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs751671453 | 18 | Y>C | No |
ExAC gnomAD |
|
| rs1348063574 | 18 | Y>H | No |
TOPMed gnomAD |
|
| rs1016604244 | 20 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1016604244 | 20 | L>V | No |
TOPMed gnomAD |
|
| rs369869047 | 22 | M>I | No |
ESP ExAC gnomAD |
|
| rs765892697 | 22 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1181389104 | 23 | L>F | No | gnomAD | |
| rs375565741 | 24 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs375565741 | 24 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1723147081 | 32 | S>G | No | Ensembl | |
| rs1237130090 | 32 | S>I | No | gnomAD | |
| rs1050007711 | 32 | S>R | No | TOPMed | |
| rs998293655 | 33 | A>T | No | TOPMed | |
| rs1447736714 | 34 | S>G | No |
TOPMed gnomAD |
|
| rs1723146689 | 34 | S>N | No | TOPMed | |
| rs374822297 | 35 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1723146455 | 36 | T>A | No | Ensembl | |
|
rs2306715 VAR_019731 |
37 | G>V | No |
UniProt ExAC TOPMed dbSNP gnomAD |
|
| rs774061111 | 38 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs1289668510 | 39 | A>G | No | gnomAD | |
| rs1289668510 | 39 | A>V | No | gnomAD | |
| rs1161206109 | 40 | A>T | No |
TOPMed gnomAD |
|
| rs1578222158 | 41 | Q>P | No | Ensembl | |
| rs1296420030 | 42 | D>E | No | gnomAD | |
| rs772445268 | 43 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs772445268 | 43 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs1743307029 | 44 | D>E | No | Ensembl | |
| rs748324562 | 45 | F>I | No |
ExAC gnomAD |
|
| rs1743306580 | 46 | F>S | No | Ensembl | |
| TCGA novel | 47 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1743306226 | 47 | H>R | No | TOPMed | |
| rs1167891231 | 48 | E>* | No | TOPMed | |
| rs1743305432 | 51 | E>A | No | TOPMed | |
| rs755367266 | 52 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs866242970 | 54 | P>L | No | gnomAD | |
| rs866242970 | 54 | P>R | No | gnomAD | |
|
rs754219253 COSM4469631 |
54 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 55 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1743303980 | 57 | P>L | No | Ensembl | |
| rs1375105736 | 58 | P>R | No |
TOPMed gnomAD |
|
| rs1363470473 | 59 | E>D | No |
TOPMed gnomAD |
|
| rs1433099464 | 61 | L>P | No | gnomAD | |
| rs1743303131 | 62 | P>L | No | TOPMed | |
| rs1743302877 | 63 | H>R | No | gnomAD | |
| rs577722147 | 66 | I>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs577722147 | 66 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1743302464 | 67 | E>G | No | TOPMed | |
| rs751105484 | 69 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs556734066 | 70 | E>D | No | Ensembl | |
| rs2149421103 | 70 | E>K | No | Ensembl | |
| rs1743301946 | 71 | A>T | No |
TOPMed gnomAD |
|
| rs1038442074 | 71 | A>V | No | Ensembl | |
| rs763542169 | 73 | I>V | No |
ExAC gnomAD |
|
| rs182706377 | 74 | V>L | No | 1000Genomes | |
| COSM4126831 | 75 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1343944791 | 76 | N>D | No | gnomAD | |
| rs1743300745 | 80 | N>I | No | Ensembl | |
|
COSM198918 rs1743300554 |
80 | N>K | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs1743300221 | 81 | L>Q | No | TOPMed | |
| rs1243208629 | 81 | L>V | No | TOPMed | |
| TCGA novel | 84 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 86 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1743299659 | 89 | T>A | No | Ensembl | |
| rs1743299284 | 90 | Q>H | No | TOPMed | |
| rs759306174 | 91 | I>M | No |
ExAC gnomAD |
|
| rs1743298889 | 92 | Y>C | No | TOPMed | |
| rs1743298748 | 96 | N>S | No | TOPMed | |
| rs4699423 | 97 | S>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1743298240 | 98 | E>K | No | TOPMed | |
| rs1743297911 | 100 | V>D | No | gnomAD | |
| rs1168795783 | 100 | V>I | No |
TOPMed gnomAD |
|
| TCGA novel | 100 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770813909 | 101 | H>L | No |
ExAC gnomAD |
|
| rs372537063 | 102 | Q>* | No |
ESP ExAC gnomAD |
|
| rs1026968329 | 102 | Q>H | No | Ensembl | |
| rs1743297387 | 102 | Q>R | No | Ensembl | |
| rs1050160011 | 104 | D>E | No | TOPMed | |
| TCGA novel | 104 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3607166 | 105 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772213131 | 106 | I>R | No |
ExAC TOPMed gnomAD |
|
| rs772213131 | 106 | I>T | No |
ExAC TOPMed gnomAD |
|
|
COSM6101124 rs774497336 COSM588840 |
107 | V>A | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs41275687 | 107 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs41275687 | 107 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs931922530 | 108 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1560791902 | 109 | E>Q | No | Ensembl | |
| rs1019905570 | 110 | R>G | No | Ensembl | |
| rs1743295901 | 110 | R>K | No |
TOPMed gnomAD |
|
| rs1743295901 | 110 | R>T | No |
TOPMed gnomAD |
|
| rs1425528620 | 111 | V>G | No | TOPMed | |
| rs535556662 | 111 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs535556662 | 111 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs780431259 | 112 | D>V | No |
ExAC gnomAD |
|
| COSM735250 | 113 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1743294928 | 115 | S>A | No | Ensembl | |
| rs1743294822 | 115 | S>C | No | Ensembl | |
| rs368269934 | 116 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 116 | G>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 117 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1388499870 | 118 | I>M | No |
TOPMed gnomAD |
|
| rs746224479 | 118 | I>T | No |
ExAC gnomAD |
|
| rs781704992 | 119 | V>A | No |
ExAC gnomAD |
|
| rs781704992 | 119 | V>G | No |
ExAC gnomAD |
|
| rs951126177 | 119 | V>I | No |
TOPMed gnomAD |
|
| rs1471595875 | 120 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
COSM1744692 rs747651633 |
120 | R>W | biliary_tract [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs751479655 | 122 | V>A | No |
ExAC gnomAD |
|
| rs1441416491 | 122 | V>L | No |
TOPMed gnomAD |
|
| rs753464899 | 123 | S>T | No |
ExAC gnomAD |
|
| rs766126524 | 124 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs750223714 | 125 | E>G | No |
1000Genomes ExAC gnomAD |
|
| rs756032843 | 125 | E>K | No |
ExAC TOPMed gnomAD |
|
|
rs575164657 COSM3607165 |
127 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes NCI-TCGA gnomAD |
| rs767605519 | 128 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| COSM6168228 | 128 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs761805868 | 128 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2149404103 | 129 | Q>R | No | Ensembl | |
| rs774320530 | 130 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1196772897 | 131 | V>A | No |
TOPMed gnomAD |
|
| rs763087403 | 131 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1346363517 | 132 | E>D | No | gnomAD | |
| rs367741337 | 134 | L>F | No |
ESP TOPMed gnomAD |
|
| COSM448366 | 136 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1469054466 | 137 | P>R | No |
TOPMed gnomAD |
|
| rs1403098822 | 138 | E>K | No | gnomAD | |
| rs769987298 | 139 | D>N | No | ExAC | |
| rs1553961921 | 143 | Q>H | No | TOPMed | |
| rs764061054 | 144 | C>R | No | Ensembl | |
| COSM735252 | 147 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759940495 | 147 | W>R | No |
ExAC gnomAD |
|
| rs776947636 | 148 | S>N | No |
ExAC gnomAD |
|
|
TCGA novel rs1188110945 |
148 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 149 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1741884670 | 149 | S>Y | No | TOPMed | |
| rs149754110 | 150 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs566430971 | 150 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs748735330 | 151 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1741883604 | 152 | T>S | No | Ensembl | |
| rs370458167 | 153 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1741883078 | 153 | T>K | No | Ensembl | |
| rs370458167 | 153 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| COSM3776086 | 155 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4126830 rs781088734 |
156 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs750278628 | 156 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs757160274 | 157 | K>R | No |
ExAC gnomAD |
|
| rs879048432 | 158 | A>S | No | Ensembl | |
|
rs751443366 COSM1671151 |
158 | A>V | central_nervous_system [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1346820880 | 159 | Y>S | No | gnomAD | |
| rs1741881837 | 160 | V>A | No | gnomAD | |
|
rs1741881583 COSM4739353 |
161 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| rs759713475 | 161 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs759713475 | 161 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs777002456 | 162 | I>L | No |
ExAC gnomAD |
|
| rs771218278 | 162 | I>T | No |
ExAC gnomAD |
|
| rs777002456 | 162 | I>V | No |
ExAC gnomAD |
|
| rs775149744 | 166 | R>L | No |
ExAC gnomAD |
|
| rs775149744 | 166 | R>Q | No |
ExAC gnomAD |
|
| rs762175329 | 166 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 167 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769358889 | 168 | T>A | No |
ExAC gnomAD |
|
| rs1454377617 | 168 | T>I | No |
TOPMed gnomAD |
|
|
rs911037326 COSM136423 |
170 | E>K | skin [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1319830598 | 171 | Q>R | No |
TOPMed gnomAD |
|
| rs1390231155 | 174 | L>P | No | gnomAD | |
| COSM3607164 | 175 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs985672419 | 177 | E>D | No |
TOPMed gnomAD |
|
| COSM73203 | 177 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1161458762 | 178 | V>A | No |
TOPMed gnomAD |
|
| rs200667278 | 181 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs200667278 | 181 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs770470612 | 182 | Q>H | No |
ExAC TOPMed gnomAD |
|
|
COSM1694701 rs1163442193 |
183 | E>K | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs2149394155 | 184 | V>I | No | Ensembl | |
| rs1446846682 | 187 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM3607163 | 188 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1421413819 | 189 | R>* | No |
TOPMed gnomAD |
|
| rs868243397 | 189 | R>Q | No |
TOPMed gnomAD |
|
| rs1740932134 | 190 | P>S | No | Ensembl | |
| TCGA novel | 192 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs777393565 COSM448365 |
193 | G>A | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA gnomAD |
| rs1415203064 | 193 | G>W | No |
TOPMed gnomAD |
|
| COSM1310396 | 195 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1215905464 | 196 | V>E | No | gnomAD | |
| COSM286322 | 197 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 198 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1579290448 | 198 | E>G | No | Ensembl | |
| COSM1310395 | 198 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779946524 | 203 | K>I | No |
ExAC gnomAD |
|
| rs923995597 | 203 | K>Q | No | Ensembl | |
| TCGA novel | 204 | N>M | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1249709216 | 206 | D>G | No |
TOPMed gnomAD |
|
| rs1739668033 | 206 | D>H | No | gnomAD | |
| rs1460812360 | 207 | I>K | No |
TOPMed gnomAD |
|
| rs373052449 | 207 | I>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs757758421 | 207 | I>M | No |
ExAC gnomAD |
|
| rs373052449 | 207 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1171703429 | 210 | P>S | No |
TOPMed gnomAD |
|
| rs1171703429 | 210 | P>T | No |
TOPMed gnomAD |
|
| rs2149382016 | 211 | V>A | No | Ensembl | |
| rs763516412 | 211 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs763516412 | 211 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1201028919 | 213 | D>N | No | gnomAD | |
| rs1490394649 | 213 | D>V | No | gnomAD | |
| COSM1059099 | 213 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs368780249 | 214 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs368780249 | 214 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs753413609 COSM4399430 |
214 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
|
TCGA novel rs1739665362 |
216 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs760380426 | 216 | F>Y | No |
ExAC gnomAD |
|
| rs376951042 | 217 | Y>C | No |
ESP ExAC |
|
| TCGA novel | 217 | Y>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs17854093 | 219 | T>I | No |
TOPMed gnomAD |
|
| rs17854093 | 219 | T>S | No |
TOPMed gnomAD |
|
| rs201855361 | 220 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1739664419 | 221 | D>E | No | Ensembl | |
| rs1739664297 | 224 | L>P | No | gnomAD | |
| rs774090101 | 230 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs141868880 | 230 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1346340329 | 231 | L>H | No | gnomAD | |
| rs749118567 | 232 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM287173 | 235 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1332692470 | 235 | A>V | No |
TOPMed gnomAD |
|
| rs1579267497 | 238 | T>P | No | Ensembl | |
| TCGA novel | 241 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 242 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1299166594 | 244 | I>F | No | gnomAD | |
| rs1299166594 | 244 | I>V | No | gnomAD | |
|
TCGA novel rs1579267469 |
246 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| TCGA novel | 248 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781321325 | 249 | K>R | No |
ExAC gnomAD |
|
| COSM4126829 | 250 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757525223 | 250 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs752006431 | 251 | T>K | No |
ExAC gnomAD |
|
| rs76956519 | 252 | T>P | No |
ExAC gnomAD |
|
| rs1230847753 | 253 | A>V | No |
TOPMed gnomAD |
|
| rs1739659918 | 254 | T>I | No | TOPMed | |
| rs753080058 | 254 | T>S | No |
ExAC gnomAD |
|
| COSM735253 | 255 | V>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1459876421 | 255 | V>I | No | gnomAD | |
| rs1272816389 | 256 | I>L | No | Ensembl | |
| rs1739659031 | 256 | I>T | No |
TOPMed gnomAD |
|
| rs1205582252 | 258 | Y>H | No |
TOPMed gnomAD |
|
| rs1205582252 | 258 | Y>N | No |
TOPMed gnomAD |
|
| rs1326477791 | 260 | N>D | No | gnomAD | |
| rs751275432 | 261 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs763719706 | 264 | S>A | No |
ExAC gnomAD |
|
| rs777573924 | 265 | T>P | No |
ExAC gnomAD |
|
| rs777573924 | 265 | T>S | No |
ExAC gnomAD |
|
| rs1579262776 | 266 | W>R | No | Ensembl | |
| rs146344947 | 267 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1660007 rs146344947 |
267 | T>M | kidney [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs146344947 | 267 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1739456570 | 268 | E>A | No | gnomAD | |
| rs1579262754 | 269 | W>G | No | Ensembl | |
| rs1739456341 | 269 | W>S | No | gnomAD | |
| rs1169545614 | 270 | S>A | No | gnomAD | |
| COSM3918385 | 270 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1169545614 | 270 | S>P | No | gnomAD | |
| rs1428399644 | 272 | C>Y | No | TOPMed | |
| rs776864880 | 273 | N>D | No |
ExAC gnomAD |
|
| rs1033493589 | 273 | N>K | No | TOPMed | |
| rs1579262727 | 273 | N>T | No | Ensembl | |
| rs776864880 | 273 | N>Y | No |
ExAC gnomAD |
|
| rs1198805080 | 274 | S>C | No | gnomAD | |
| rs1739455036 | 275 | R>C | No | TOPMed | |
| rs771070280 | 275 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM1541010 | 277 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1417647452 COSM242180 |
278 | R>* | Variant assessed as Somatic; HIGH impact. pancreas large_intestine prostate [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
| rs138978640 | 278 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs138978640 | 278 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1324353084 | 279 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1579262696 | 279 | G>R | No | Ensembl | |
| rs1579262682 | 280 | Y>D | No | Ensembl | |
| rs1437584015 | 283 | R>C | No | gnomAD | |
|
rs201199826 COSM3607162 |
283 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1437584015 | 283 | R>S | No | gnomAD | |
| rs748492018 | 284 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs748492018 | 284 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs1739453587 | 286 | T>I | No | Ensembl | |
| rs1282027763 | 287 | C>S | No | gnomAD | |
| TCGA novel | 290 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs559427223 | 290 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs559427223 | 290 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756813685 | 291 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs763779157 | 293 | L>F | No |
ExAC gnomAD |
|
| rs763779157 | 293 | L>V | No |
ExAC gnomAD |
|
| rs758033265 | 294 | N>S | No |
ExAC TOPMed gnomAD |
|
|
rs1428521407 COSM3661279 |
295 | G>E | liver [Cosmic] | No |
cosmic curated gnomAD |
| rs1220854873 | 297 | A>D | No | TOPMed | |
| rs1175748210 | 298 | F>L | No | gnomAD | |
| rs2149379603 | 299 | C>S | No | 1000Genomes | |
| rs2149379600 | 302 | Q>K | No | Ensembl | |
| rs1162923313 | 303 | S>R | No | gnomAD | |
| COSM448364 | 303 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1739451578 | 304 | V>M | No | Ensembl | |
|
COSM4126828 rs894307669 |
307 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs776778306 | 307 | I>T | No |
ExAC gnomAD |
|
| rs1473139408 | 307 | I>V | No | gnomAD | |
| rs1207084026 | 308 | A>S | No | gnomAD | |
| rs1739450862 | 308 | A>V | No | Ensembl | |
| rs1263096561 | 309 | C>F | No | gnomAD | |
| rs1201179493 | 310 | T>I | No | gnomAD | |
| rs1201179493 | 310 | T>S | No | gnomAD | |
| rs200437262 | 311 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs200437262 | 311 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1472589668 | 312 | L>F | No | gnomAD | |
| rs2149379573 | 312 | L>S | No | Ensembl | |
| rs1222836353 | 313 | C>R | No | gnomAD | |
| TCGA novel | 314 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754860601 | 316 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs1430145819 | 316 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1420587009 | 317 | G>R | No | gnomAD | |
| rs529568856 | 318 | R>S | No |
1000Genomes ExAC gnomAD |
|
| rs1579260820 | 319 | W>G | No | Ensembl | |
| rs766450239 | 320 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1739365690 | 322 | W>C | No | TOPMed | |
| rs935612399 | 324 | K>E | No |
TOPMed gnomAD |
|
| TCGA novel | 324 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM735255 | 325 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1579260785 | 325 | W>G | No | Ensembl | |
| rs1243382118 | 326 | S>T | No | gnomAD | |
| COSM3918384 | 329 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1739364417 | 332 | C>S | No | TOPMed | |
| rs1739364292 | 332 | C>Y | No | gnomAD | |
| rs200814959 | 333 | T>N | No |
1000Genomes ExAC gnomAD |
|
| rs200814959 | 333 | T>S | No |
1000Genomes ExAC gnomAD |
|
| rs933030381 | 334 | H>R | No | gnomAD | |
| rs1294801129 | 335 | W>S | No |
TOPMed gnomAD |
|
| rs768734972 | 336 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs763344659 | 336 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs768734972 | 336 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1739363019 | 338 | R>G | No | TOPMed | |
| rs1340050600 | 338 | R>S | No | TOPMed | |
| rs1300190593 | 338 | R>T | No |
TOPMed gnomAD |
|
| rs1400777010 | 339 | E>G | No | gnomAD | |
| COSM3607160 | 339 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1579260710 | 340 | C>G | No | Ensembl | |
|
rs775796452 COSM4739352 |
341 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs775796452 | 341 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs777156310 | 342 | A>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs777156310 COSM1431886 |
342 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs778676302 | 343 | P>A | No |
ExAC gnomAD |
|
| rs754647343 | 343 | P>L | No |
ExAC gnomAD |
|
| rs925152623 | 344 | A>P | No | Ensembl | |
| rs925152623 | 344 | A>T | No | Ensembl | |
| rs1425137759 | 345 | P>L | No | gnomAD | |
| COSM3918383 | 345 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs865857865 | 346 | K>E | No | Ensembl | |
|
COSM1231802 rs2149378441 |
346 | K>N | large_intestine [Cosmic] | No |
cosmic curated Ensembl |
| rs1193775931 | 348 | G>R | No | gnomAD | |
| TCGA novel | 350 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753707650 | 351 | D>G | No |
ExAC gnomAD |
|
| rs1739360804 | 351 | D>Y | No | Ensembl | |
| rs201363473 | 353 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1250589067 | 353 | D>G | No | gnomAD | |
| rs145155041 | 353 | D>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145155041 | 353 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM1059097 rs1243736071 |
354 | G>S | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
| rs1739359538 | 355 | L>F | No |
TOPMed gnomAD |
|
| COSM6168230 | 355 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1739359020 | 356 | V>D | No | TOPMed | |
|
rs371874775 COSM1328336 |
356 | V>I | ovary Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| COSM1059096 | 357 | L>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1348986751 | 359 | S>P | No | TOPMed | |
| rs1435183834 | 359 | S>Y | No |
TOPMed gnomAD |
|
| rs759650789 | 360 | K>Q | No |
TOPMed gnomAD |
|
| rs1739358366 | 360 | K>R | No | Ensembl | |
| rs1345065125 | 362 | C>* | No | gnomAD | |
| rs1018638106 | 364 | D>H | No |
TOPMed gnomAD |
|
| COSM6101125 | 364 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM273698 | 364 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1739357864 | 365 | G>R | No | Ensembl | |
| rs1391896003 | 366 | L>F | No |
TOPMed gnomAD |
|
| rs1739357636 | 366 | L>H | No | Ensembl | |
| rs1391896003 | 366 | L>I | No |
TOPMed gnomAD |
|
| rs1227490958 | 368 | M>T | No | TOPMed | |
| rs1443056480 | 371 | A>V | No |
TOPMed gnomAD |
|
| TCGA novel | 374 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2149368324 | 375 | D>H | No | Ensembl | |
| rs1738418707 | 377 | V>A | No | Ensembl | |
| rs1356195603 | 377 | V>I | No |
TOPMed gnomAD |
|
| rs558173942 | 378 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748790797 | 379 | L>F | No |
ExAC gnomAD |
|
| rs1286445334 | 379 | L>P | No | gnomAD | |
| rs748790797 | 379 | L>V | No |
ExAC gnomAD |
|
| rs769538762 | 380 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs769538762 | 380 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs745704670 | 381 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs745704670 | 381 | V>D | No |
ExAC TOPMed gnomAD |
|
| rs745704670 | 381 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs866262514 | 382 | G>V | No | Ensembl | |
| rs1401398180 | 383 | I>V | No | gnomAD | |
| rs1231711017 | 384 | V>A | No |
TOPMed gnomAD |
|
| rs780804804 | 385 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs757235410 | 386 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs757235410 | 386 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1738416267 | 386 | A>V | No | TOPMed | |
| rs777686939 | 387 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1163744061 | 388 | I>T | No |
TOPMed gnomAD |
|
|
rs752752057 COSM1059094 |
389 | V>I | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs765485658 COSM2989140 |
392 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs766583475 | 395 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 395 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1450204038 | 396 | V>I | No | gnomAD | |
| rs200326223 | 397 | V>A | No |
1000Genomes gnomAD |
|
| rs1738414297 | 397 | V>L | No | TOPMed | |
| rs1292554024 | 398 | A>T | No | gnomAD | |
| rs773913128 | 399 | L>S | No |
ExAC gnomAD |
|
| rs747586731 | 402 | Y>* | No |
ExAC gnomAD |
|
| rs762588223 | 403 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs762588223 | 403 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs200067795 | 403 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1282221999 | 405 | N>D | No | gnomAD | |
| rs1402765519 | 406 | H>Q | No | gnomAD | |
| rs1424888543 | 406 | H>R | No | gnomAD | |
|
COSM448363 rs138355335 |
407 | R>C | breast [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs150089453 | 407 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1738412233 | 408 | D>N | No | gnomAD | |
| rs1738411845 | 409 | F>S | No |
TOPMed gnomAD |
|
| rs776491788 | 411 | S>A | No |
ExAC TOPMed |
|
| rs776491788 | 411 | S>T | No |
ExAC TOPMed |
|
| rs770701663 | 412 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs1478829968 | 412 | D>E | No |
TOPMed gnomAD |
|
| rs770701663 | 412 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1216430505 | 412 | D>N | No | TOPMed | |
| rs377428633 | 413 | I>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs377428633 | 413 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs375351610 | 414 | I>V | No |
ESP TOPMed gnomAD |
|
| rs1579241996 | 416 | S>Y | No | Ensembl | |
|
COSM4438007 rs202087388 |
417 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1738409552 | 418 | A>P | No |
TOPMed gnomAD |
|
| rs753021614 | 420 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs753021614 | 420 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs753021614 | 420 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs1256744960 | 421 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1440685948 | 422 | G>V | No |
TOPMed gnomAD |
|
| rs550835877 | 424 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1005066859 | 424 | Q>R | No |
TOPMed gnomAD |
|
| rs2149368186 | 425 | P>H | No | Ensembl | |
| rs756484272 | 428 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1738408012 | 429 | K>E | No | TOPMed | |
| rs1217103081 | 429 | K>N | No |
TOPMed gnomAD |
|
| COSM1495882 | 430 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1369302469 | 431 | A>V | No |
TOPMed gnomAD |
|
| rs186748241 | 432 | R>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs186748241 | 432 | R>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1441249743 | 433 | Q>* | No | gnomAD | |
|
rs376083297 COSM3940987 |
435 | L>M | oesophagus [Cosmic] | No |
cosmic curated ESP TOPMed gnomAD |
| rs376083297 | 435 | L>V | No |
ESP TOPMed gnomAD |
|
| rs1160839442 | 436 | L>R | No | gnomAD | |
| rs1738377902 | 438 | V>I | No | Ensembl | |
| rs1738377466 | 439 | P>H | No | gnomAD | |
| rs1243105943 | 439 | P>S | No |
TOPMed gnomAD |
|
| rs1243105943 | 439 | P>T | No |
TOPMed gnomAD |
|
| rs1738377346 | 440 | P>A | No | gnomAD | |
| rs1738376933 | 442 | L>F | No | Ensembl | |
| rs140743706 | 443 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs140743706 | 443 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs757848056 | 445 | A>T | No |
ExAC gnomAD |
|
| rs1378309866 | 445 | A>V | No | TOPMed | |
| COSM4126826 | 447 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1224161269 | 448 | M>K | No |
TOPMed gnomAD |
|
| rs1224161269 | 448 | M>T | No |
TOPMed gnomAD |
|
| rs752182179 | 449 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1738375637 | 450 | R>K | No |
TOPMed gnomAD |
|
| rs1738375515 | 452 | P>S | No | Ensembl | |
| rs539639757 | 454 | Y>C | No |
1000Genomes ExAC gnomAD |
|
| rs759021473 | 455 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs760273584 | 458 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1303560244 | 458 | D>G | No |
TOPMed gnomAD |
|
| rs961401548 | 459 | V>A | No |
TOPMed gnomAD |
|
| rs767365642 | 459 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs767365642 | 459 | V>L | No |
ExAC TOPMed gnomAD |
|
|
COSM588843 COSM6101126 rs761581481 |
461 | D>E | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1738373692 | 462 | K>E | No | gnomAD | |
| TCGA novel | 462 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774203936 | 464 | P>S | No |
ExAC gnomAD |
|
| rs774203936 | 464 | P>T | No |
ExAC gnomAD |
|
| rs768606046 | 465 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs768606046 | 465 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs571344977 | 466 | T>P | No | 1000Genomes | |
|
rs147587723 COSM4126825 |
467 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| COSM1059093 | 469 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1185431885 | 470 | I>V | No | gnomAD | |
| rs1008338067 | 472 | D>Y | No | Ensembl | |
| rs568825787 | 473 | P>Q | No |
1000Genomes ExAC |
|
| rs537865725 | 473 | P>S | No |
1000Genomes ExAC gnomAD |
|
| rs537865725 | 473 | P>T | No |
1000Genomes ExAC gnomAD |
|
| rs74479359 | 474 | L>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1738371694 | 475 | P>S | No | TOPMed | |
| TCGA novel | 476 | N>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 479 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747359888 | 481 | V>M | No |
ExAC gnomAD |
|
|
COSM198902 rs1738370835 |
482 | Y>C | large_intestine [Cosmic] | No |
cosmic curated Ensembl |
| rs778184879 | 482 | Y>H | No |
ExAC gnomAD |
|
| TCGA novel | 484 | T>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765858771 | 487 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs755615649 | 487 | A>V | No |
ExAC gnomAD |
|
| rs529245772 | 488 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1169277663 | 489 | T>I | No | gnomAD | |
| rs1351004233 | 489 | T>P | No | gnomAD | |
| rs763956322 | 491 | Q>E | No |
ExAC gnomAD |
|
| rs763956322 | 491 | Q>K | No |
ExAC gnomAD |
|
| TCGA novel | 491 | Q>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 491 | Q>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775433817 | 493 | D>E | No |
ExAC TOPMed gnomAD |
|
| COSM6101128 | 493 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1264935265 | 494 | L>F | No | gnomAD | |
| rs745932278 | 495 | S>P | No |
ExAC gnomAD |
|
| rs1579240986 | 496 | E>Q | No | Ensembl | |
| rs771250502 | 498 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1738367681 | 499 | S>C | No | TOPMed | |
| rs1260138439 | 499 | S>T | No | gnomAD | |
| rs1579240956 | 500 | K>T | No | Ensembl | |
| rs758844127 | 501 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs758844127 | 501 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1358529032 | 502 | S>C | No | TOPMed | |
| rs1358529032 | 502 | S>F | No | TOPMed | |
| rs1442763889 | 503 | P>S | No | TOPMed | |
| rs779272894 | 504 | Q>H | No | ExAC | |
| rs1236790002 | 504 | Q>K | No | gnomAD | |
|
TCGA novel rs1388906091 |
505 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
gnomAD NCI-TCGA |
| rs1299589013 | 505 | M>K | No |
TOPMed gnomAD |
|
| rs1299589013 | 505 | M>R | No |
TOPMed gnomAD |
|
| rs1299589013 | 505 | M>T | No |
TOPMed gnomAD |
|
| rs1738365997 | 506 | T>N | No | Ensembl | |
| rs755668812 | 506 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs2149367668 | 507 | Q>H | No | 1000Genomes | |
| rs1738365681 | 507 | Q>L | No | TOPMed | |
| rs1464583952 | 508 | S>L | No | gnomAD | |
| rs1738365426 | 509 | L>M | No | TOPMed | |
| rs760152083 | 510 | L>S | No |
ExAC gnomAD |
|
| rs760152083 | 510 | L>W | No |
ExAC gnomAD |
|
| rs373018154 | 511 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1465176942 | 511 | E>G | No | gnomAD | |
| rs756850221 | 511 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs762655060 | 513 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs762655060 | 513 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs765247673 | 514 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs752741406 | 514 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs765247673 | 514 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs759735193 | 516 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1738363692 | 516 | S>R | No | TOPMed | |
| rs760894608 | 517 | L>R | No |
ExAC gnomAD |
|
| rs771301532 | 517 | L>V | No |
ExAC gnomAD |
|
| rs1344516387 | 518 | K>R | No | gnomAD | |
| rs1235745512 | 519 | N>D | No | gnomAD | |
| rs949113007 | 519 | N>K | No |
1000Genomes gnomAD |
|
| COSM175781 | 519 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200139331 | 520 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1738362152 | 521 | S>N | No | TOPMed | |
| rs1738361974 | 521 | S>R | No | gnomAD | |
| rs1379773409 | 521 | S>R | No | gnomAD | |
| rs920459275 | 522 | L>V | No |
TOPMed gnomAD |
|
| rs779435582 | 524 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs1738361360 | 526 | T>A | No | Ensembl | |
| rs201518489 | 526 | T>I | No |
TOPMed gnomAD |
|
| rs201518489 | 526 | T>S | No |
TOPMed gnomAD |
|
| rs200661813 | 527 | D>A | No | Ensembl | |
| rs1738360734 | 528 | P>S | No | Ensembl | |
| COSM368175 | 528 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2149367584 | 529 | S>Y | No | Ensembl | |
| rs1410040469 | 530 | C>F | No | TOPMed | |
| rs369513609 | 530 | C>W | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM2989122 rs148691835 |
532 | A>T | Variant assessed as Somatic; MODERATE impact. pancreas [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1303477323 | 533 | F>L | No |
TOPMed gnomAD |
|
| rs530815653 | 533 | F>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777280898 | 533 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs375565874 | 534 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1738359040 | 535 | S>R | No | TOPMed | |
| rs1738358902 | 537 | N>D | No | Ensembl | |
| rs1274480862 | 537 | N>S | No |
TOPMed gnomAD |
|
| TCGA novel | 537 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 538 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs919830330 COSM4126824 |
538 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs765300766 | 538 | S>T | No |
ExAC TOPMed gnomAD |
|
| COSM3607158 | 540 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753980631 | 543 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1738357784 | 544 | I>V | No | TOPMed | |
| rs766500731 | 546 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs766500731 | 546 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1238474243 | 548 | S>A | No | gnomAD | |
|
rs1471182317 COSM6101129 |
549 | G>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| COSM4126823 | 551 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1738243195 | 551 | S>T | No | Ensembl | |
| rs1157387685 | 554 | I>L | No |
TOPMed gnomAD |
|
| rs767802168 | 554 | I>M | No |
ExAC TOPMed gnomAD |
|
| COSM3607157 | 555 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 555 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1197030 rs774698598 |
556 | A>S | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs774698598 | 556 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM1541012 | 556 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 558 | A>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 558 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763287953 | 560 | P>S | No |
ExAC gnomAD |
|
| COSM3607155 | 562 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1738241800 | 562 | G>R | No | TOPMed | |
| rs746563619 | 564 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs746563619 | 564 | V>G | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 564 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747737623 | 566 | E>G | No |
ExAC gnomAD |
|
| rs185464975 | 566 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs754880404 | 567 | M>I | No |
ExAC gnomAD |
|
| TCGA novel | 568 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1738240369 | 570 | T>S | No | gnomAD | |
| rs1335888740 | 572 | H>Q | No |
TOPMed gnomAD |
|
| rs1220027112 | 572 | H>R | No | gnomAD | |
| rs201871465 | 572 | H>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2149366406 | 573 | R>G | No | Ensembl | |
|
COSM3607154 rs1219708513 |
573 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1738239412 | 574 | K>E | No |
TOPMed gnomAD |
|
| rs759903005 | 579 | P>T | No |
ExAC gnomAD |
|
| rs1263098208 | 580 | P>L | No | Ensembl | |
| rs1457720153 | 580 | P>S | No | gnomAD | |
| rs1553953281 | 581 | M>V | No | Ensembl | |
| rs1737894075 | 584 | S>F | No | TOPMed | |
| rs1200019054 | 585 | Q>H | No | gnomAD | |
| rs1737893674 | 586 | T>A | No | TOPMed | |
| rs1277451656 | 589 | T>I | No | gnomAD | |
| COSM735259 | 590 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1737893256 | 591 | V>L | No | gnomAD | |
| rs1737893013 | 592 | V>A | No | gnomAD | |
| COSM4126822 | 593 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs747650161 | 593 | S>N | No |
ExAC gnomAD |
|
| rs1024527050 | 594 | C>Y | No | gnomAD | |
| rs779656103 | 596 | P>R | No |
ExAC gnomAD |
|
| rs749116875 | 596 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs749116875 | 596 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs756078264 | 598 | G>E | No |
ExAC gnomAD |
|
|
COSM6168232 COSM1541013 rs2149362395 |
601 | L>F | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic Ensembl |
| rs1355225600 | 601 | L>P | No | gnomAD | |
| rs1737891761 | 602 | T>N | No | gnomAD | |
|
rs139568380 COSM3409633 |
603 | R>C | central_nervous_system [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs376193832 | 603 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA gnomAD |
| rs1737891015 | 604 | P>Q | No | TOPMed | |
| rs1181110178 | 605 | V>A | No | gnomAD | |
|
COSM5571687 rs987321967 |
605 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
|
rs777197285 COSM4126821 |
606 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs2149362364 | 607 | L>H | No | Ensembl | |
| rs1737889695 | 608 | T>I | No | Ensembl | |
| COSM402493 | 609 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs370003428 | 609 | M>L | No |
ESP TOPMed gnomAD |
|
| rs765458013 | 609 | M>T | No |
ExAC gnomAD |
|
| rs370003428 | 609 | M>V | No |
ESP TOPMed gnomAD |
|
| rs759944822 | 610 | H>P | No |
ExAC TOPMed gnomAD |
|
| rs759944822 | 610 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs900538848 | 611 | H>Q | No | TOPMed | |
| rs761436712 | 613 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs761436712 | 613 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM1158674 rs761436712 |
613 | A>T | pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs768433445 | 614 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1737888308 | 615 | P>S | No | Ensembl | |
| rs762623193 | 616 | N>D | No |
ExAC gnomAD |
|
| rs762623193 | 616 | N>H | No |
ExAC gnomAD |
|
| rs1377535766 | 617 | T>I | No |
TOPMed gnomAD |
|
| rs1359071747 | 618 | E>G | No | gnomAD | |
| rs745651856 | 618 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1488312392 | 619 | D>N | No | gnomAD | |
| TCGA novel | 621 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1180391405 | 623 | L>P | No |
TOPMed gnomAD |
|
| rs200871650 | 626 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs770817442 | 627 | Q>R | No |
ExAC gnomAD |
|
| rs141976218 | 628 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146123204 | 628 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1257750447 | 629 | A>T | No | gnomAD | |
| TCGA novel | 629 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs150755514 | 630 | Q>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150755514 | 630 | Q>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs752932977 | 632 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs989886706 | 633 | W>C | No | TOPMed | |
| rs755349239 | 633 | W>R | No |
ExAC gnomAD |
|
| rs1169344969 | 635 | D>V | No |
TOPMed gnomAD |
|
| rs938986045 | 636 | V>M | No |
TOPMed gnomAD |
|
| rs1480813025 | 637 | V>M | No |
TOPMed gnomAD |
|
| rs1432356553 | 640 | G>E | No |
TOPMed gnomAD |
|
| rs776415886 | 640 | G>R | No |
ExAC TOPMed gnomAD |
|
| COSM4126820 | 640 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1737740886 | 641 | E>G | No | TOPMed | |
| rs1265086741 | 641 | E>Q | No |
TOPMed gnomAD |
|
| COSM3918382 | 642 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1195412727 | 643 | N>I | No |
TOPMed gnomAD |
|
| rs1195412727 | 643 | N>S | No |
TOPMed gnomAD |
|
| rs1468872887 | 644 | F>L | No |
TOPMed gnomAD |
|
| rs1253635927 | 644 | F>S | No | gnomAD | |
| rs1025531104 | 645 | T>P | No | gnomAD | |
| rs149451558 | 646 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs149451558 | 646 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1013710374 | 646 | T>P | No | gnomAD | |
| rs773186313 | 647 | P>R | No |
ExAC gnomAD |
|
| rs760453427 | 647 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1737738990 | 649 | Y>H | No | gnomAD | |
| rs1579227518 | 652 | L>R | No | Ensembl | |
| rs772018154 | 653 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs1737738374 | 654 | A>E | No | Ensembl | |
| rs558984414 | 655 | E>G | No |
1000Genomes ExAC gnomAD |
|
| rs1737738251 | 655 | E>K | No | TOPMed | |
| rs768761666 | 656 | A>D | No |
ExAC gnomAD |
|
| rs774427913 | 656 | A>P | No |
ExAC gnomAD |
|
| rs774427913 | 656 | A>T | No |
ExAC gnomAD |
|
| TCGA novel | 657 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1305810151 COSM3661275 |
659 | I>N | liver [Cosmic] | No |
cosmic curated Ensembl |
| rs1369479901 | 661 | T>K | No | TOPMed | |
| rs756595601 | 662 | E>D | No |
ExAC gnomAD |
|
| rs952354401 | 662 | E>Q | No | Ensembl | |
| rs1737736668 | 663 | N>I | No | Ensembl | |
| rs1737736556 | 663 | N>K | No | Ensembl | |
| rs1737736413 | 664 | L>F | No | gnomAD | |
| rs746202995 | 665 | S>C | No |
ExAC gnomAD |
|
| rs746202995 | 665 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1454631910 | 665 | S>N | No | gnomAD | |
| rs1440524220 | 666 | T>P | No | Ensembl | |
| rs1737735832 | 667 | Y>C | No | TOPMed | |
| rs187196396 | 668 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1417395006 COSM1059088 |
668 | A>V | Variant assessed as Somatic; MODERATE impact. skin endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1195756746 | 672 | H>L | No |
TOPMed gnomAD |
|
| rs2149360674 | 673 | S>A | No | Ensembl | |
| rs201608163 | 673 | S>F | No |
1000Genomes TOPMed gnomAD |
|
| rs201608163 | 673 | S>Y | No |
1000Genomes TOPMed gnomAD |
|
| rs1737734189 | 674 | T>I | No | TOPMed | |
| rs374566244 | 675 | T>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs374566244 | 675 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1737733626 | 676 | K>N | No |
TOPMed gnomAD |
|
| rs754442498 | 677 | A>E | No |
ExAC TOPMed gnomAD |
|
| COSM278012 | 677 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs754442498 COSM286321 |
677 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1337002863 | 678 | A>S | No |
TOPMed gnomAD |
|
| rs1337002863 | 678 | A>T | No |
TOPMed gnomAD |
|
| rs1307103039 | 679 | A>T | No |
TOPMed gnomAD |
|
| rs61736724 | 679 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs141250673 | 681 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM165406 rs774148486 |
681 | R>H | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs768834811 | 685 | A>T | No |
ExAC gnomAD |
|
| rs1389195192 | 687 | F>C | No |
TOPMed gnomAD |
|
| rs763103301 | 687 | F>I | No |
ExAC TOPMed gnomAD |
|
| rs763103301 | 687 | F>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 688 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs117896462 | 689 | P>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs117896462 | 689 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs117896462 | 689 | P>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1423412479 | 690 | L>P | No |
TOPMed gnomAD |
|
| rs1423412479 | 690 | L>R | No |
TOPMed gnomAD |
|
| rs1042964643 | 691 | C>R | No | Ensembl | |
| rs1190382253 | 694 | S>L | No |
TOPMed gnomAD |
|
| rs2149360613 | 698 | S>G | No | Ensembl | |
| rs1737728845 | 698 | S>N | No | TOPMed | |
|
TCGA novel rs1737728731 |
699 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
|
COSM227480 rs771347599 |
700 | R>Q | Variant assessed as Somatic; MODERATE impact. endometrium skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| COSM448362 | 701 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM6101131 | 704 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 706 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754568367 | 707 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs754568367 | 707 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs754568367 | 707 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1560730281 | 708 | Q>P | No | Ensembl | |
| COSM2989115 | 709 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779715793 | 710 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs753318607 | 710 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs779715793 | 710 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1737727165 | 712 | K>E | No | TOPMed | |
| rs1243737398 | 713 | E>D | No |
TOPMed gnomAD |
|
| rs779468452 | 714 | I>S | No |
ExAC TOPMed gnomAD |
|
| rs779468452 | 714 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1386963353 | 716 | H>D | No | gnomAD | |
| rs2062156591 | 716 | H>Q | No | Ensembl | |
| COSM1633855 | 716 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1301046271 | 717 | L>F | No |
TOPMed gnomAD |
|
| rs1579208937 | 718 | E>D | No | TOPMed | |
| rs563357325 | 719 | R>T | No |
1000Genomes ExAC gnomAD |
|
| rs1181571366 | 720 | Q>H | No | TOPMed | |
| rs151009512 | 721 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
|
rs2289043 VAR_019732 |
721 | M>T | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1392307583 | 721 | M>V | No | gnomAD | |
| rs1736782354 | 722 | G>A | No | gnomAD | |
| rs2149351648 | 722 | G>R | No | Ensembl | |
| rs757114910 | 723 | G>R | No |
ExAC gnomAD |
|
| rs1175723434 | 724 | Q>E | No |
TOPMed gnomAD |
|
| rs1476578500 | 727 | E>A | No | gnomAD | |
| rs751272161 | 727 | E>K | No |
ExAC gnomAD |
|
| rs751272161 | 727 | E>Q | No |
ExAC gnomAD |
|
| rs1579208870 | 728 | E>D | No | Ensembl | |
| rs574421593 | 729 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1374048071 | 729 | P>T | No |
TOPMed gnomAD |
|
| COSM1059087 | 733 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779554924 | 734 | F>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 736 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 736 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765481264 | 737 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1270410976 | 738 | T>A | No |
TOPMed gnomAD |
|
| rs776961360 | 740 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs1339815696 | 741 | L>M | No |
TOPMed gnomAD |
|
|
rs1736779087 COSM5498893 |
742 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
|
rs142722408 COSM5649681 |
742 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs112874916 | 743 | L>P | No | Ensembl | |
| TCGA novel | 743 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761134744 | 746 | H>D | No |
ExAC gnomAD |
|
| rs773402815 | 746 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs761134744 | 746 | H>Y | No |
ExAC gnomAD |
|
| rs146792764 | 747 | D>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs139502011 | 747 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146792764 | 747 | D>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs143643587 | 748 | I>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs769339200 | 749 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs745338820 | 749 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1397571731 | 753 | W>C | No | gnomAD | |
| rs1248751319 | 755 | S>C | No | TOPMed | |
| rs933309029 | 755 | S>R | No |
TOPMed gnomAD |
|
| rs1172390007 | 756 | K>I | No | gnomAD | |
| rs1172390007 | 756 | K>R | No | gnomAD | |
| rs1397402100 | 757 | L>F | No | gnomAD | |
| rs1453062318 | 757 | L>V | No | gnomAD | |
| rs1194848389 | 758 | L>P | No | gnomAD | |
| rs1194848389 | 758 | L>R | No | gnomAD | |
| rs1736775665 | 760 | K>T | No | TOPMed | |
| rs780928372 | 762 | Q>E | No |
ExAC gnomAD |
|
| TCGA novel | 763 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs769110355 COSM137410 |
763 | E>K | skin [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs769110355 | 763 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs775944665 | 764 | I>T | No |
ExAC gnomAD |
|
| rs1413229804 | 765 | P>R | No | gnomAD | |
| rs1357693858 | 767 | Y>H | No |
TOPMed gnomAD |
|
| TCGA novel | 767 | Y>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1736681471 | 768 | H>Y | No | TOPMed | |
| rs1313875409 | 769 | V>I | No |
TOPMed gnomAD |
|
| rs1736681243 | 771 | S>C | No | Ensembl | |
| rs1343972810 | 772 | G>* | No | gnomAD | |
| rs746707534 | 772 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs1343972810 | 772 | G>R | No | gnomAD | |
| rs1370112761 | 775 | R>T | No | gnomAD | |
| TCGA novel | 776 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 776 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1736679960 | 778 | H>Y | No | TOPMed | |
|
COSM1059086 rs758318256 |
780 | T>I | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1736679462 | 781 | F>I | No | TOPMed | |
| rs747921696 | 781 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs1243743837 | 782 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs779028369 | 783 | L>M | No |
ExAC gnomAD |
|
| rs377562086 | 784 | E>G | No |
ESP ExAC gnomAD |
|
| rs1337265834 | 785 | R>G | No | gnomAD | |
| rs1360366123 | 786 | F>C | No | gnomAD | |
| rs1381989934 | 786 | F>L | No |
TOPMed gnomAD |
|
| rs1736677904 | 787 | S>G | No | Ensembl | |
| rs756203732 | 788 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs756203732 | 788 | L>Q | No |
ExAC TOPMed gnomAD |
|
| rs766433438 | 788 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs767692546 | 789 | N>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 790 | T>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751872601 | 791 | V>A | No |
ExAC gnomAD |
|
| rs1426511017 | 792 | E>K | No | gnomAD | |
| rs1190124765 | 793 | L>M | No |
TOPMed gnomAD |
|
| TCGA novel | 793 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201410096 | 796 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 799 | V>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs773039868 COSM3768066 |
800 | R>Q | liver [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs547179134 | 800 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1469918990 | 802 | V>G | No |
TOPMed gnomAD |
|
| rs1274011930 | 803 | E>A | No | gnomAD | |
| rs1274011930 | 803 | E>G | No | gnomAD | |
| rs1180387773 | 805 | E>G | No |
TOPMed gnomAD |
|
|
COSM3918381 rs927398355 |
806 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs747983287 | 808 | I>T | No |
ExAC gnomAD |
|
| rs1579206676 | 809 | F>L | No | Ensembl | |
| rs768646708 | 810 | Q>* | No |
ExAC gnomAD |
|
| rs1301943389 | 810 | Q>R | No |
TOPMed gnomAD |
|
| COSM735260 | 811 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs144306140 | 812 | N>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs74806677 | 812 | N>S | No | Ensembl | |
| rs144306140 | 812 | N>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs780017284 | 813 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs756184035 | 814 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs139372477 | 815 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1431880 rs139372477 |
815 | V>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1195468947 | 817 | E>A | No | gnomAD | |
| rs1213619060 | 817 | E>K | No |
TOPMed gnomAD |
|
| rs1213619060 | 817 | E>Q | No |
TOPMed gnomAD |
|
| rs773054467 | 818 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs778034359 | 819 | P>L | No |
ExAC TOPMed |
|
| rs2149343408 | 819 | P>T | No | Ensembl | |
| rs150900256 | 821 | G>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs150900256 | 821 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1736043547 | 822 | I>V | No | gnomAD | |
| rs1352180970 | 823 | D>E | No |
TOPMed gnomAD |
|
| rs761855500 | 823 | D>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 824 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs143332038 | 825 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs773911528 | 827 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1560710613 | 828 | D>H | No | Ensembl | |
| rs373876244 | 829 | P>S | No |
ESP TOPMed gnomAD |
|
| rs763759984 | 830 | A>P | No |
ExAC gnomAD |
|
| rs1160105752 | 830 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1736041096 | 831 | N>T | No | TOPMed | |
| rs1052136157 | 832 | T>A | No | TOPMed | |
| rs1022635500 | 832 | T>S | No |
TOPMed gnomAD |
|
| rs1736040545 | 833 | I>N | No | TOPMed | |
| rs1190539989 | 834 | T>A | No | gnomAD | |
| rs796889648 | 834 | T>I | No | TOPMed | |
| rs745871673 | 836 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs745871673 | 836 | V>D | No |
ExAC TOPMed gnomAD |
|
| rs1203963050 | 836 | V>F | No | gnomAD | |
| rs1203963050 | 836 | V>L | No | gnomAD | |
| rs368284839 | 837 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs368284839 | 837 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 838 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1466296291 | 840 | S>R | No |
TOPMed gnomAD |
|
| rs142730145 | 840 | S>T | No |
ESP TOPMed |
|
|
rs34585936 VAR_055327 |
841 | A>T | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1233657813 | 842 | F>I | No | gnomAD | |
| rs779272234 | 843 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs754442812 | 845 | P>L | No |
ExAC TOPMed |
|
| rs755394328 | 845 | P>S | No | ExAC | |
| rs929930790 | 846 | L>P | No | gnomAD | |
| rs1383857833 | 847 | P>S | No |
TOPMed gnomAD |
|
| rs756857276 | 848 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs756857276 | 848 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs751028526 | 849 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM2989102 rs897744414 |
849 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs1198848015 | 850 | Q>L | No |
TOPMed gnomAD |
|
| rs762597746 | 851 | K>N | No |
ExAC gnomAD |
|
| rs1736035665 | 852 | L>H | No | TOPMed | |
| rs1255019194 | 853 | C>G | No | Ensembl | |
| TCGA novel | 854 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1167434160 | 854 | S>N | No | gnomAD | |
| rs1473321989 | 854 | S>R | No | gnomAD | |
| rs775000272 | 855 | S>C | No |
ExAC gnomAD |
|
| rs765074261 | 856 | L>R | No |
ExAC gnomAD |
|
| TCGA novel | 858 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1259484834 | 858 | A>V | No | TOPMed | |
| rs776591203 | 859 | P>L | No |
ExAC gnomAD |
|
| rs372767649 | 860 | Q>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs773288996 | 860 | Q>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 860 | Q>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201959747 | 861 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs755431038 | 862 | R>K | No |
ExAC gnomAD |
|
| rs946762918 | 863 | G>C | No |
TOPMed gnomAD |
|
| rs946762918 | 863 | G>S | No |
TOPMed gnomAD |
|
| rs749626575 | 866 | W>* | No |
ExAC gnomAD |
|
| rs749626575 | 866 | W>C | No |
ExAC gnomAD |
|
| rs1736032837 | 867 | R>G | No | Ensembl | |
| TCGA novel | 867 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756628543 | 868 | M>I | No |
ExAC gnomAD |
|
| rs780726697 | 868 | M>T | No |
ExAC gnomAD |
|
|
rs145024545 COSM390228 |
871 | H>N | lung [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs145024545 | 871 | H>Y | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 876 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 876 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757832625 | 877 | R>G | No |
ExAC gnomAD |
|
| COSM5002449 | 877 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1183608637 | 878 | Y>F | No | gnomAD | |
| rs2149342949 | 879 | L>F | No | Ensembl | |
| TCGA novel | 881 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1735995640 | 882 | F>L | No | TOPMed | |
| rs752109152 | 882 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs752109152 | 882 | F>Y | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 884 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1735994891 | 886 | S>C | No |
TOPMed gnomAD |
|
| TCGA novel | 886 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1219851648 | 889 | T>I | No |
TOPMed gnomAD |
|
| rs1219851648 | 889 | T>N | No |
TOPMed gnomAD |
|
| rs1280773317 | 889 | T>P | No | gnomAD | |
| rs1219851648 | 889 | T>S | No |
TOPMed gnomAD |
|
| rs1350125407 | 890 | G>S | No | gnomAD | |
| rs199997284 | 891 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1735993472 | 892 | I>V | No | Ensembl | |
| rs750426754 | 894 | D>N | No | ExAC | |
| rs750426754 | 894 | D>Y | No | ExAC | |
| rs980020057 | 900 | N>H | No |
TOPMed gnomAD |
|
| TCGA novel | 900 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1735992589 | 900 | N>S | No |
TOPMed gnomAD |
|
| rs1319092340 | 901 | F>V | No |
TOPMed gnomAD |
|
| rs1177486467 | 902 | P>A | No |
TOPMed gnomAD |
|
| rs1177486467 | 902 | P>S | No |
TOPMed gnomAD |
|
| rs777195597 | 903 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1735991389 | 904 | G>E | No | TOPMed | |
| rs775839267 | 904 | G>R | No |
ExAC gnomAD |
|
| rs746186375 | 908 | M>I | No |
ExAC gnomAD |
|
| rs2149342897 | 908 | M>V | No | Ensembl | |
| rs2149342892 | 909 | L>P | No | Ensembl | |
| rs573022437 | 911 | A>P | No |
1000Genomes ExAC gnomAD |
|
| rs747709581 | 913 | L>F | No |
ExAC gnomAD |
|
| rs1735990017 | 915 | E>G | No | TOPMed | |
| rs1282267112 | 916 | M>I | No |
TOPMed gnomAD |
|
| rs1275713270 | 917 | G>A | No |
TOPMed gnomAD |
|
|
TCGA novel rs1275713270 |
917 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs778329637 | 917 | G>R | No |
ExAC gnomAD |
|
| rs754484155 | 918 | R>K | No |
ExAC TOPMed |
|
| rs754484155 | 918 | R>T | No |
ExAC TOPMed |
|
| rs1735989284 | 920 | E>K | No | Ensembl | |
| rs1735989094 | 921 | T>A | No | gnomAD | |
| rs749000531 | 921 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs755980772 | 922 | V>L | No |
ExAC gnomAD |
|
| rs2149342868 | 923 | V>A | No | Ensembl | |
| rs553218717 | 923 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767501437 | 924 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs2149342860 | 925 | L>S | No | Ensembl | |
| TCGA novel | 926 | A>E | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1035123993 | 926 | A>T | No | TOPMed | |
| rs751536528 | 927 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs1192678046 | 927 | A>S | No |
TOPMed gnomAD |
|
| rs751536528 | 927 | A>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 928 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764325623 | 928 | E>K | No |
ExAC gnomAD |
|
| rs763103180 | 929 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs765338334 | 931 | Y>H | No |
ExAC gnomAD |
|
| rs1049058813 | 932 | Y>L | No | TOPMed | |
| rs1735986301 | 932 | Y>Q | No | Ensembl | |
| rs1049058813 | 932 | Y>S | No | TOPMed | |
| rs1414269035 | 932 | Y>S | No | gnomAD | |
| rs1735985529 | 932 | Y>Y | No | TOPMed |
No associated diseases with O95185
15 regional properties for O95185
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 621 - 882 | IPR000719 |
| domain | Ephrin receptor ligand binding domain | 30 - 209 | IPR001090 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 621 - 878 | IPR001245 |
| conserved_site | Tyrosine-protein kinase, receptor class V, conserved site | 185 - 205 | IPR001426-1 |
| conserved_site | Tyrosine-protein kinase, receptor class V, conserved site | 247 - 267 | IPR001426-2 |
| domain | Sterile alpha motif domain | 908 - 975 | IPR001660 |
| domain | Fibronectin type III | 328 - 439 | IPR003961-1 |
| domain | Fibronectin type III | 440 - 537 | IPR003961-2 |
| active_site | Tyrosine-protein kinase, active site | 742 - 754 | IPR008266 |
| domain | Tyrosine-protein kinase ephrin type A/B receptor-like | 268 - 306 | IPR011641 |
| binding_site | Protein kinase, ATP binding site | 627 - 653 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 621 - 878 | IPR020635 |
| domain | Ephrin receptor, transmembrane domain | 550 - 618 | IPR027936 |
| domain | Ephrin type-A receptor 4, SAM domain | 911 - 981 | IPR030602 |
| domain | Ephrin type-A receptor 4, ligand binding domain | 30 - 203 | IPR034270 |
Functions
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell surface | The external part of the cell wall and/or plasma membrane. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| growth cone | The migrating motile tip of a growing neuron projection, where actin accumulates, and the actin cytoskeleton is the most dynamic. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| netrin receptor activity | Combining with a netrin signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity. |
| netrin receptor activity involved in chemorepulsion | Combining with a netrin signal and transmitting the signal from one side of the membrane to the other to contribute to the directed movement of a motile cell away from a higher concentration of netrin. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| tubulin binding | Binding to monomeric or multimeric forms of tubulin, including microtubules. |
11 GO annotations of biological process
| Name | Definition |
|---|---|
| anterior/posterior axon guidance | The process in which the migration of an axon growth cone is directed to a specific target site along the anterior-posterior body axis in response to a combination of attractive and repulsive cues. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism. |
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| chemorepulsion of axon | The process in which a neuron growth cone is directed to a specific target site in response to a repulsive chemical cue. |
| dorsal root ganglion development | The process whose specific outcome is the progression of a dorsal root ganglion over time, from its formation to the mature structure. |
| ectopic germ cell programmed cell death | Programmed cell death of an errant germ line cell that is outside the normal migratory path or ectopic to the gonad. This is an important mechanism of regulating germ cell survival within the embryo. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of developmental process | Any process that activates or increases the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult). |
| positive regulation of reproductive process | Any process that activates or increases the frequency, rate or extent of reproductive process. |
| regulation of neuron migration | Any process that modulates the frequency, rate or extent of neuron migration. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q7T2Z5 | UNC5C | Netrin receptor UNC5C | Gallus gallus (Chicken) | SS |
| Q6ZN44 | UNC5A | Netrin receptor UNC5A | Homo sapiens (Human) | SS |
| Q8IZJ1 | UNC5B | Netrin receptor UNC5B | Homo sapiens (Human) | SS |
| Q6UXZ4 | UNC5D | Netrin receptor UNC5D | Homo sapiens (Human) | SS |
| Q8K1S2 | Unc5d | Netrin receptor UNC5D | Mus musculus (Mouse) | SS |
| Q8K1S3 | Unc5b | Netrin receptor UNC5B | Mus musculus (Mouse) | SS |
| Q8K1S4 | Unc5a | Netrin receptor UNC5A | Mus musculus (Mouse) | SS |
| O08747 | Unc5c | Netrin receptor UNC5C | Mus musculus (Mouse) | SS |
| F1LW30 | Unc5d | Netrin receptor UNC5D | Rattus norvegicus (Rat) | SS |
| O08721 | Unc5a | Netrin receptor UNC5A | Rattus norvegicus (Rat) | SS |
| O08722 | Unc5b | Netrin receptor UNC5B | Rattus norvegicus (Rat) | EV |
| Q761X5 | Unc5c | Netrin receptor UNC5C | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRKGLRATAA | RCGLGLGYLL | QMLVLPALAL | LSASGTGSAA | QDDDFFHELP | ETFPSDPPEP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LPHFLIEPEE | AYIVKNKPVN | LYCKASPATQ | IYFKCNSEWV | HQKDHIVDER | VDETSGLIVR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EVSIEISRQQ | VEELFGPEDY | WCQCVAWSSA | GTTKSRKAYV | RIAYLRKTFE | QEPLGKEVSL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EQEVLLQCRP | PEGIPVAEVE | WLKNEDIIDP | VEDRNFYITI | DHNLIIKQAR | LSDTANYTCV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AKNIVAKRKS | TTATVIVYVN | GGWSTWTEWS | VCNSRCGRGY | QKRTRTCTNP | APLNGGAFCE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GQSVQKIACT | TLCPVDGRWT | PWSKWSTCGT | ECTHWRRREC | TAPAPKNGGK | DCDGLVLQSK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| NCTDGLCMQT | APDSDDVALY | VGIVIAVIVC | LAISVVVALF | VYRKNHRDFE | SDIIDSSALN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GGFQPVNIKA | ARQDLLAVPP | DLTSAAAMYR | GPVYALHDVS | DKIPMTNSPI | LDPLPNLKIK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VYNTSGAVTP | QDDLSEFTSK | LSPQMTQSLL | ENEALSLKNQ | SLARQTDPSC | TAFGSFNSLG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GHLIVPNSGV | SLLIPAGAIP | QGRVYEMYVT | VHRKETMRPP | MDDSQTLLTP | VVSCGPPGAL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LTRPVVLTMH | HCADPNTEDW | KILLKNQAAQ | GQWEDVVVVG | EENFTTPCYI | QLDAEACHIL |
| 670 | 680 | 690 | 700 | 710 | 720 |
| TENLSTYALV | GHSTTKAAAK | RLKLAIFGPL | CCSSLEYSIR | VYCLDDTQDA | LKEILHLERQ |
| 730 | 740 | 750 | 760 | 770 | 780 |
| MGGQLLEEPK | ALHFKGSTHN | LRLSIHDIAH | SLWKSKLLAK | YQEIPFYHVW | SGSQRNLHCT |
| 790 | 800 | 810 | 820 | 830 | 840 |
| FTLERFSLNT | VELVCKLCVR | QVEGEGQIFQ | LNCTVSEEPT | GIDLPLLDPA | NTITTVTGPS |
| 850 | 860 | 870 | 880 | 890 | 900 |
| AFSIPLPIRQ | KLCSSLDAPQ | TRGHDWRMLA | HKLNLDRYLN | YFATKSSPTG | VILDLWEAQN |
| 910 | 920 | 930 | |||
| FPDGNLSMLA | AVLEEMGRHE | TVVSLAAEGQ | Y |