O94768
Gene name |
STK17B (DRAK2) |
Protein name |
Serine/threonine-protein kinase 17B |
Names |
DAP kinase-related apoptosis-inducing protein kinase 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9262 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
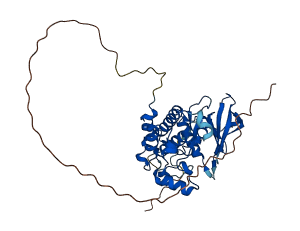
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
178-200 (Activation loop from InterPro)
Target domain |
33-293 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

11 structures for O94768
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3LM0 | X-ray | 235 A | A | 25-329 | PDB |
| 3LM5 | X-ray | 229 A | A | 25-329 | PDB |
| 6QF4 | X-ray | 250 A | A | 18-329 | PDB |
| 6Y6F | X-ray | 198 A | A | 25-329 | PDB |
| 6Y6H | X-ray | 195 A | A | 25-329 | PDB |
| 6ZJF | X-ray | 175 A | A | 25-329 | PDB |
| 7AKG | X-ray | 208 A | A | 25-329 | PDB |
| 7Q7C | X-ray | 285 A | A | 25-329 | PDB |
| 7Q7D | X-ray | 260 A | A | 25-329 | PDB |
| 7Q7E | X-ray | 285 A | A | 25-329 | PDB |
| AF-O94768-F1 | Predicted | AlphaFoldDB |
225 variants for O94768
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA2037433 rs779126465 |
2 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs1461579485 CA350011955 |
3 | R>K | No |
ClinGen TOPMed |
|
|
rs1270955712 CA350011948 |
4 | R>K | No |
ClinGen gnomAD |
|
|
CA350011943 rs1220160297 |
5 | R>G | No |
ClinGen gnomAD |
|
|
rs950185817 CA63491679 |
6 | F>V | No |
ClinGen Ensembl |
|
|
rs917529164 CA63491678 |
7 | D>G | No |
ClinGen Ensembl |
|
|
rs983698538 CA63491677 |
9 | R>* | No |
ClinGen TOPMed gnomAD |
|
|
CA2037430 rs375355841 |
9 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA2037429 rs766362335 |
11 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA63491675 rs202160682 |
15 | L>P | No |
ClinGen 1000Genomes gnomAD |
|
|
CA2037426 rs767393117 |
17 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2037425 rs761357694 |
18 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA63491674 rs878907400 |
24 | K>* | No |
ClinGen Ensembl |
|
|
CA350011813 rs1252700707 |
25 | M>I | No |
ClinGen TOPMed |
|
|
rs1227596869 CA350011818 |
25 | M>V | No |
ClinGen TOPMed |
|
|
CA350011806 rs1179852701 |
26 | E>G | No |
ClinGen gnomAD |
|
|
CA350011809 rs1320886016 |
26 | E>Q | No |
ClinGen TOPMed |
|
|
CA63491673 rs962476081 |
27 | N>K | No |
ClinGen Ensembl |
|
|
CA2037423 rs754831402 TCGA novel |
28 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ExAC gnomAD NCI-TCGA |
|
rs753775218 CA63491671 |
29 | N>K | No |
ClinGen Ensembl |
|
|
rs1252581057 CA350011785 |
29 | N>S | No |
ClinGen gnomAD |
|
|
CA350011767 rs1202649425 |
31 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA2037422 rs763740195 |
33 | I>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 35 | T>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA350011734 rs1211352653 |
36 | S>F | No |
ClinGen gnomAD |
|
|
CA350011732 rs1266791246 |
37 | K>E | No |
ClinGen gnomAD |
|
|
rs761281792 CA2037418 |
38 | E>K | No |
ClinGen ExAC |
|
|
CA63491669 rs1006706069 |
39 | L>R | No |
ClinGen TOPMed |
|
|
rs1455229887 CA350011686 |
41 | R>S | No |
ClinGen TOPMed |
|
|
rs1345451941 CA350011676 |
43 | K>R | No |
ClinGen TOPMed |
|
|
CA2037391 rs781126284 |
44 | F>S | No |
ClinGen ExAC gnomAD |
|
|
rs1370195092 CA350011652 |
47 | V>I | No |
ClinGen gnomAD |
|
|
rs1164482277 CA350011646 |
48 | R>G | No |
ClinGen gnomAD |
|
|
rs1351414204 CA350011643 |
48 | R>T | No |
ClinGen gnomAD |
|
|
CA350011637 rs1449036129 |
49 | Q>* | No |
ClinGen gnomAD |
|
|
rs751311997 CA2037389 |
49 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1447933258 CA350011629 |
50 | C>Y | No |
ClinGen gnomAD |
|
|
CA2037387 rs777580805 |
51 | I>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs750899409 CA63490916 |
54 | S>F | No |
ClinGen Ensembl |
|
|
rs200641057 CA2037385 |
55 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA350011584 rs1246047751 |
57 | Q>R | No |
ClinGen gnomAD |
|
| TCGA novel | 58 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs764651841 CA2037383 |
58 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs752455132 CA2037384 |
58 | E>Q | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 58 | E>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1300144690 CA350011569 |
59 | Y>* | No |
ClinGen gnomAD |
|
|
CA63490915 rs555479339 |
59 | Y>C | No |
ClinGen Ensembl |
|
|
rs759023174 CA2037382 |
61 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA63490914 rs867537241 |
63 | F>S | No |
ClinGen Ensembl |
|
|
CA63490913 rs537143112 |
65 | K>R | No |
ClinGen TOPMed |
|
|
rs934597790 CA63490912 |
66 | K>T | No |
ClinGen Ensembl |
|
| TCGA novel | 67 | R>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA63490911 rs144100898 COSM1136552 |
69 | R>G | kidney [Cosmic] | No |
ClinGen cosmic curated 1000Genomes gnomAD |
|
CA350011499 rs1241154913 |
70 | G>E | No |
ClinGen Ensembl |
|
|
rs762441909 CA2037379 |
72 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs774826980 CA2037378 |
74 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
COSM718700 rs190316595 CA2037377 |
74 | R>Q | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed |
|
rs1283595338 CA350011470 |
75 | A>T | No |
ClinGen TOPMed |
|
|
CA63490909 rs867130560 |
78 | L>S | No |
ClinGen Ensembl |
|
|
rs769921646 CA2037374 |
81 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA350011418 rs1270198295 |
82 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
CA2037373 rs745907345 |
82 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA2037372 rs374612008 |
83 | V>A | No |
ClinGen ESP ExAC TOPMed |
|
|
CA350011385 rs1482437365 |
87 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1271007800 CA350011384 |
88 | K>E | No |
ClinGen TOPMed |
|
|
rs746994801 CA2037370 |
89 | S>C | No |
ClinGen ExAC |
|
|
rs551545854 CA63490907 |
91 | P>L | No |
ClinGen gnomAD |
|
|
CA350011361 rs1559413717 |
91 | P>S | No |
ClinGen Ensembl |
|
|
rs370398613 CA2037369 |
92 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs758258757 CA2037368 |
92 | R>H | No |
ClinGen ExAC gnomAD |
|
|
CA350011311 rs1180855668 |
99 | V>I | No |
ClinGen TOPMed |
|
|
CA350011280 rs1283844693 |
103 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA350011276 rs1409349861 |
103 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1409349861 CA350011277 |
103 | T>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1368713731 CA350011274 |
104 | S>R | No |
ClinGen gnomAD |
|
|
rs376276095 CA2037367 |
107 | I>V | No |
ClinGen ESP ExAC |
|
|
rs1333027449 CA350011180 |
114 | A>P | No |
ClinGen TOPMed |
|
|
CA350011176 rs1414802791 |
114 | A>V | No |
ClinGen gnomAD |
|
| TCGA novel | 116 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA63489690 rs373607600 |
118 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA350011132 rs1222420056 |
121 | L>V | No |
ClinGen gnomAD |
|
|
rs1340688049 CA350011110 |
124 | P>R | No |
ClinGen TOPMed |
|
|
CA350011103 rs1236595079 |
125 | E>G | No |
ClinGen gnomAD |
|
|
CA350011070 rs1184884487 CA350011071 |
129 | M>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs779704372 CA2037345 |
134 | D>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 135 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs951453448 CA63489689 |
136 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
CA350011025 rs1207856731 |
136 | I>T | No |
ClinGen gnomAD |
|
|
CA2037344 rs755616780 |
136 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1286259477 CA350011023 |
137 | R>G | No |
ClinGen gnomAD |
|
|
rs1218516775 CA350011020 |
137 | R>K | No |
ClinGen TOPMed |
|
|
CA350011018 rs1575174528 |
137 | R>S | No |
ClinGen Ensembl |
|
|
CA2037342 rs764646680 |
139 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1282652553 CA350011001 |
140 | K>E | No |
ClinGen gnomAD |
|
|
rs1450414760 CA350010998 |
140 | K>R | No |
ClinGen gnomAD |
|
| TCGA novel | 143 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1320359791 CA350010978 |
143 | L>V | No |
ClinGen gnomAD |
|
|
CA2037341 rs758856381 |
146 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA2037340 rs753070528 |
148 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1341487554 CA350010935 |
149 | L>R | No |
ClinGen TOPMed |
|
|
CA350010912 rs1400795784 |
152 | N>K | No |
ClinGen gnomAD |
|
|
rs1274206603 CA350010817 |
164 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA350010775 rs1465799572 |
169 | I>M | No |
ClinGen gnomAD |
|
|
CA2037312 rs749048042 |
169 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA350010756 rs1218976730 |
171 | P>S | No |
ClinGen gnomAD |
|
| TCGA novel | 173 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4139149 CA2037310 rs768648951 |
173 | G>R | ovary [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA63489460 rs894728755 |
175 | I>V | No |
ClinGen Ensembl |
|
| TCGA novel | 180 | F>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs762784446 CA2037309 |
184 | R>Q | No |
ClinGen ExAC |
|
|
CA350010501 rs1441398660 |
186 | I>M | No |
ClinGen Ensembl |
|
|
rs373533424 CA350010485 |
188 | H>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs993377576 CA63489459 |
188 | H>R | No |
ClinGen TOPMed |
|
|
CA2037308 rs373533424 |
188 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1460112130 CA350010434 |
190 | C>S | No |
ClinGen TOPMed gnomAD |
|
|
CA350010433 rs1460112130 |
190 | C>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs1057415621 CA63489457 |
193 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA2037304 rs770413196 |
196 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA350010337 rs1423043519 |
196 | M>R | No |
ClinGen gnomAD |
|
|
rs780835050 CA2037305 |
196 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA63489456 rs938912477 |
197 | G>E | No |
ClinGen Ensembl |
|
|
CA2037287 rs776077952 |
204 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1204682887 CA350013115 |
207 | L>P | No |
ClinGen gnomAD |
|
|
CA2037286 rs770632480 |
210 | D>V | No |
ClinGen ExAC gnomAD |
|
|
CA2037284 rs772810962 |
211 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA2037285 rs746456183 |
211 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 212 | I>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1395050701 CA350013071 |
214 | T>S | No |
ClinGen gnomAD |
|
|
CA2037282 rs539135098 |
217 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 217 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs765268119 CA2037270 |
221 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA350013005 rs1364105050 |
222 | G>S | No |
ClinGen gnomAD |
|
|
CA2037268 rs776508128 |
226 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs765908410 CA2037267 |
234 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs200455780 CA2037266 |
240 | N>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1195983797 CA350012839 |
246 | N>D | No |
ClinGen gnomAD |
|
|
CA2037264 rs771631226 |
246 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA350012826 rs1054537 |
247 | I>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA63489065 rs980845542 |
249 | Q>H | No |
ClinGen Ensembl |
|
|
CA63489064 rs376869706 |
253 | D>G | No |
ClinGen ESP TOPMed |
|
|
rs1185960279 CA350012790 |
253 | D>Y | No |
ClinGen TOPMed |
|
|
rs571951842 CA2037262 |
254 | Y>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA63489063 rs969582838 |
255 | S>L | No |
ClinGen TOPMed gnomAD |
|
|
CA2037260 rs746176436 |
260 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA350012730 rs1428418794 |
262 | V>F | No |
ClinGen TOPMed |
|
| TCGA novel | 263 | S>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs771032608 CA2037257 |
265 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA2037256 rs747314013 |
266 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs777709836 CA2037255 |
277 | N>K | No |
ClinGen ExAC gnomAD |
|
|
CA63489061 rs571636905 |
279 | E>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1013864396 CA63489062 |
279 | E>K | No |
ClinGen Ensembl |
|
|
rs1482933982 CA350012580 |
282 | P>L | No |
ClinGen TOPMed |
|
|
CA2037242 rs148605491 |
286 | I>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2037241 rs770251222 |
287 | C>R | No |
ClinGen ExAC gnomAD |
|
|
CA350012551 rs1303579315 |
287 | C>Y | No |
ClinGen gnomAD |
|
|
rs1392337811 CA350012521 |
291 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
CA2037240 rs760133191 |
294 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
rs776845846 CA2037239 |
297 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA350012468 rs374655630 |
298 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA2037237 rs747337215 |
300 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2037236 rs781385899 |
304 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA2037235 rs772447182 |
307 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs998674437 CA63488850 |
307 | T>N | No |
ClinGen TOPMed |
|
|
rs779175254 CA2037233 |
308 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA2037232 rs754849303 |
308 | S>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2037230 rs779713505 |
309 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 310 | S>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs755874663 CA350012383 |
311 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA2037229 rs755874663 |
311 | S>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA63488849 rs768797284 |
313 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA350012364 rs1348546526 |
314 | Q>P | No |
ClinGen gnomAD |
|
|
rs1032242724 CA63488848 |
315 | D>G | No |
ClinGen TOPMed |
|
|
CA350012360 rs1281521567 |
315 | D>H | No |
ClinGen gnomAD |
|
|
CA350012350 rs1340069852 |
316 | H>L | No |
ClinGen gnomAD |
|
|
rs750202113 CA2037228 |
316 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs113234544 CA63488846 |
318 | V>A | No |
ClinGen TOPMed |
|
|
rs1335188050 CA350012340 |
318 | V>I | No |
ClinGen gnomAD |
|
|
CA2037227 rs767114137 |
319 | R>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2037226 rs34740616 VAR_041147 |
320 | S>F | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1320406056 CA350012321 |
321 | S>C | No |
ClinGen gnomAD |
|
|
CA350012315 rs1559407263 |
322 | E>G | No |
ClinGen Ensembl |
|
|
rs751042475 CA2037225 |
323 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA350012295 rs1282535814 |
325 | T>A | No |
ClinGen TOPMed |
|
|
CA2037223 rs759955994 |
326 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs771353477 CA2037221 |
328 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1181597307 CA350012268 |
329 | S>C | No |
ClinGen gnomAD |
|
|
rs773745315 CA2037219 |
330 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA350012255 rs1214936223 |
331 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA2037218 rs117222182 |
334 | C>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA350012236 rs1286486560 |
334 | C>S | No |
ClinGen gnomAD |
|
|
CA350012237 rs1286486560 |
334 | C>Y | No |
ClinGen gnomAD |
|
|
CA2037217 rs200748046 |
335 | G>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA63488845 rs111690654 |
336 | D>G | No |
ClinGen Ensembl |
|
|
rs774560861 CA2037216 |
337 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA350012218 rs1454350777 |
337 | R>T | No |
ClinGen TOPMed |
|
|
CA2037215 rs768851113 |
338 | E>A | No |
ClinGen ExAC gnomAD |
|
|
CA2037214 rs749336239 |
339 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2037213 rs779986694 |
339 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749336239 CA350012206 |
339 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1297554181 CA350012198 |
340 | K>T | No |
ClinGen gnomAD |
|
|
CA350012193 rs1418770008 |
341 | E>K | No |
ClinGen TOPMed |
|
|
rs185557932 CA2037212 |
343 | I>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA350012171 rs1347443642 |
344 | P>S | No |
ClinGen gnomAD |
|
|
CA350012155 rs1319765530 |
346 | D>H | No |
ClinGen gnomAD |
|
|
CA2037211 rs575911954 |
347 | S>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 347 | S>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs939088383 CA63488843 |
348 | S>N | No |
ClinGen Ensembl |
|
|
rs927833723 CA63488842 |
349 | M>V | No |
ClinGen TOPMed |
|
|
CA63488841 rs972956606 |
350 | V>A | No |
ClinGen Ensembl |
|
|
rs1446255982 CA350012114 |
352 | K>E | No |
ClinGen gnomAD |
|
|
rs939941689 CA63488840 |
353 | R>I | No |
ClinGen Ensembl |
|
|
CA2037208 COSM1014363 rs756886954 |
355 | R>C | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM3709300 CA2037207 rs751279324 |
355 | R>H | liver [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA63488838 CA350012078 rs752170351 |
357 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA63488839 rs983992477 |
357 | D>G | No |
ClinGen Ensembl |
|
|
rs142101505 CA2037205 |
357 | D>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA2037203 rs766870205 |
358 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs566842605 CA2037201 |
359 | S>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA2037198 rs372377287 |
362 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2037197 rs774653030 |
363 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA350012022 rs1400094453 |
366 | L>F | No |
ClinGen gnomAD |
|
|
rs1344278902 CA350012007 |
368 | S>L | No |
ClinGen gnomAD |
|
|
CA350012006 rs1158819815 |
369 | D>H | No |
ClinGen TOPMed |
|
|
CA350011984 rs1226743030 |
372 | C>R | No |
ClinGen gnomAD |
|
|
CA2037195 rs749342673 |
373 | C>W | No |
ClinGen ExAC gnomAD |
No associated diseases with O94768
4 regional properties for O94768
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 33 - 293 | IPR000719 |
| active_site | Serine/threonine-protein kinase, active site | 154 - 166 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 39 - 62 | IPR017441 |
| domain | Serine/threonine-protein kinase 17B, catalytic domain | 24 - 293 | IPR042763 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| endoplasmic reticulum-Golgi intermediate compartment | A complex system of membrane-bounded compartments located between endoplasmic reticulum (ER) and the Golgi complex, with a distinctive membrane protein composition; involved in ER-to-Golgi and Golgi-to-ER transport. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of fibroblast apoptotic process | Any process that activates or increases the frequency, rate or extent of fibroblast apoptotic process. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
99 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4IFM7 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Bos taurus (Bovine) | SS |
| Q0V7M1 | PSKH1 | Serine/threonine-protein kinase H1 | Bos taurus (Bovine) | SS |
| Q0VD22 | STK33 | Serine/threonine-protein kinase 33 | Bos taurus (Bovine) | PR |
| Q96NX5 | CAMK1G | Calcium/calmodulin-dependent protein kinase type 1G | Homo sapiens (Human) | SS |
| O15075 | DCLK1 | Serine/threonine-protein kinase DCLK1 | Homo sapiens (Human) | EV |
| Q14012 | CAMK1 | Calcium/calmodulin-dependent protein kinase type 1 | Homo sapiens (Human) | EV |
| Q86YV6 | MYLK4 | Myosin light chain kinase family member 4 | Homo sapiens (Human) | SS |
| Q16566 | CAMK4 | Calcium/calmodulin-dependent protein kinase type IV | Homo sapiens (Human) | SS |
| P11801 | PSKH1 | Serine/threonine-protein kinase H1 | Homo sapiens (Human) | SS |
| Q8N568 | DCLK2 | Serine/threonine-protein kinase DCLK2 | Homo sapiens (Human) | PR |
| O75962 | TRIO | Triple functional domain protein | Homo sapiens (Human) | EV |
| Q9H1R3 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Homo sapiens (Human) | EV |
| Q32MK0 | MYLK3 | Myosin light chain kinase 3 | Homo sapiens (Human) | SS |
| Q8WWF8 | CAPSL | Calcyphosin-like protein | Homo sapiens (Human) | PR |
| Q6P2M8 | PNCK | Calcium/calmodulin-dependent protein kinase type 1B | Homo sapiens (Human) | SS |
| Q9BYT3 | STK33 | Serine/threonine-protein kinase 33 | Homo sapiens (Human) | PR |
| Q8NCB2 | CAMKV | CaM kinase-like vesicle-associated protein | Homo sapiens (Human) | SS |
| Q8IU85 | CAMK1D | Calcium/calmodulin-dependent protein kinase type 1D | Homo sapiens (Human) | SS |
| Q9UEE5 | STK17A | Serine/threonine-protein kinase 17A | Homo sapiens (Human) | SS |
| P53355 | DAPK1 | Death-associated protein kinase 1 | Homo sapiens (Human) | EV |
| O43293 | DAPK3 | Death-associated protein kinase 3 | Homo sapiens (Human) | PR |
| Q9UIK4 | DAPK2 | Death-associated protein kinase 2 | Homo sapiens (Human) | EV |
| Q9QYK9 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Mus musculus (Mouse) | PR |
| Q91VB2 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Mus musculus (Mouse) | SS |
| Q8VCR8 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Mus musculus (Mouse) | SS |
| Q6PGN3 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Mus musculus (Mouse) | PR |
| Q91YS8 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Mus musculus (Mouse) | SS |
| Q3UIZ8 | Mylk3 | Myosin light chain kinase 3 | Mus musculus (Mouse) | SS |
| Q8VDF3 | Dapk2 | Death-associated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q6P8Y1 | Capsl | Calcyphosin-like protein | Mus musculus (Mouse) | PR |
| Q8BW96 | Camk1d | Calcium/calmodulin-dependent protein kinase type 1D | Mus musculus (Mouse) | SS |
| Q3UHL1 | Camkv | CaM kinase-like vesicle-associated protein | Mus musculus (Mouse) | SS |
| P08414 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Mus musculus (Mouse) | SS |
| Q9JLM8 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Mus musculus (Mouse) | SS |
| Q91YA2 | Pskh1 | Serine/threonine-protein kinase H1 | Mus musculus (Mouse) | SS |
| Q80YE7 | Dapk1 | Death-associated protein kinase 1 | Mus musculus (Mouse) | SS |
| Q62407 | Speg | Striated muscle-specific serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| Q0KL02 | Trio | Triple functional domain protein | Mus musculus (Mouse) | SS |
| Q924X7 | Stk33 | Serine/threonine-protein kinase 33 | Mus musculus (Mouse) | PR |
| O54784 | Dapk3 | Death-associated protein kinase 3 | Mus musculus (Mouse) | PR |
| Q8BG48 | Stk17b | Serine/threonine-protein kinase 17B | Mus musculus (Mouse) | PR |
| Q7TNJ7 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Rattus norvegicus (Rat) | SS |
| P20689 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Rattus norvegicus (Rat) | SS |
| F1M0Z1 | Trio | Triple functional domain protein | Rattus norvegicus (Rat) | SS |
| O08875 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Rattus norvegicus (Rat) | PR |
| E9PT87 | Mylk3 | Myosin light chain kinase 3 | Rattus norvegicus (Rat) | SS |
| O70150 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Rattus norvegicus (Rat) | PR |
| Q63092 | Camkv | CaM kinase-like vesicle-associated protein | Rattus norvegicus (Rat) | SS |
| Q63638 | Speg | Striated muscle-specific serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| O88764 | Dapk3 | Death-associated protein kinase 3 | Rattus norvegicus (Rat) | PR |
| P13234 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Rattus norvegicus (Rat) | SS |
| Q63450 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Rattus norvegicus (Rat) | EV |
| P97924 | Kalrn | Kalirin | Rattus norvegicus (Rat) | SS |
| Q5MPA9 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Rattus norvegicus (Rat) | PR |
| Q91XS8 | Stk17b | Serine/threonine-protein kinase 17B | Rattus norvegicus (Rat) | PR |
| Q5VQQ5 | CPK2 | Calcium-dependent protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q8LPZ7 | CPK3 | Calcium-dependent protein kinase 3 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6K968 | CPK6 | Calcium-dependent protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| P53684 | CPK7 | Calcium-dependent protein kinase 7 | Oryza sativa subsp japonica (Rice) | PR |
| Q75GE8 | CPK8 | Calcium-dependent protein kinase 8 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6I5I8 | CPK16 | Calcium-dependent protein kinase 16 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6AVM3 | CCAMK | Calcium and calcium/calmodulin-dependent serine/threonine-protein kinase | Oryza sativa subsp japonica (Rice) | PR |
| Q84SL0 | CPK20 | Calcium-dependent protein kinase 20 | Oryza sativa subsp. japonica (Rice) | SS |
| P53682 | CPK23 | Calcium-dependent protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| Q9TXJ0 | cmk-1 | Calcium/calmodulin-dependent protein kinase type 1 | Caenorhabditis elegans | PR |
| O44997 | dapk-1 | Death-associated protein kinase dapk-1 | Caenorhabditis elegans | SS |
| Q95QC4 | zyg-8 | Serine/threonine-protein kinase zyg-8 | Caenorhabditis elegans | PR |
| P28583 | Calcium-dependent protein kinase SK5 | Glycine max (Soybean) (Glycine hispida) | PR | |
| Q9M9V8 | CPK10 | Calcium-dependent protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39016 | CPK11 | Calcium-dependent protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FX86 | CRK8 | CDPK-related kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SSF8 | CPK30 | Calcium-dependent protein kinase 30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38872 | CPK6 | Calcium-dependent protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7XJR9 | CPK16 | Calcium-dependent protein kinase 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIQ7 | CPK24 | Calcium-dependent protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJ61 | CPK25 | Calcium-dependent protein kinase 25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZV15 | CPK20 | Calcium-dependent protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93759 | GK-1 | Calcium-dependent protein kinase 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8S8S2 | LPEAT2 | Lysophospholipid acyltransferase LPEAT2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38870 | CPK2 | Calcium-dependent protein kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LJL9 | CRK2 | CDPK-related kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SG12 | CRK6 | CDPK-related kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCS2 | CRK5 | CDPK-related kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W4I7 | CPK13 | Calcium-dependent protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NLQ6 | CPK32 | Calcium-dependent protein kinase 32 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZSA4 | CPK27 | Calcium-dependent protein kinase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38869 | CPK4 | Calcium-dependent protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42479 | CPK3 | Calcium-dependent protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1PE17 | CPK18 | Calcium-dependent protein kinase 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q06850 | CPK1 | Calcium-dependent protein kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FMP5 | CPK17 | Calcium-dependent protein kinase 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38873 | CPK7 | Calcium-dependent protein kinase 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E9C0 | CPK34 | Calcium-dependent protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42438 | CPK8 | Calcium-dependent protein kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42396 | CPK12 | Calcium-dependent protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FKW4 | CPK28 | Calcium-dependent protein kinase 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1LUA6 | trio | Triple functional domain protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A8C984 | mylk3 | Myosin light chain kinase 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q7SY49 | camkv | CaM kinase-like vesicle-associated protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSRRRFDCRS | ISGLLTTTPQ | IPIKMENFNN | FYILTSKELG | RGKFAVVRQC | ISKSTGQEYA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AKFLKKRRRG | QDCRAEILHE | IAVLELAKSC | PRVINLHEVY | ENTSEIILIL | EYAAGGEIFS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LCLPELAEMV | SENDVIRLIK | QILEGVYYLH | QNNIVHLDLK | PQNILLSSIY | PLGDIKIVDF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GMSRKIGHAC | ELREIMGTPE | YLAPEILNYD | PITTATDMWN | IGIIAYMLLT | HTSPFVGEDN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QETYLNISQV | NVDYSEETFS | SVSQLATDFI | QSLLVKNPEK | RPTAEICLSH | SWLQQWDFEN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LFHPEETSSS | SQTQDHSVRS | SEDKTSKSSC | NGTCGDREDK | ENIPEDSSMV | SKRFRFDDSL |
| 370 | |||||
| PNPHELVSDL | LC |