O88705
Gene name |
Hcn3 (Hac3) |
Protein name |
Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 |
Names |
Hyperpolarization-activated cation channel 3 , HAC-3 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:15168 |
EC number |
|
Protein Class |
|
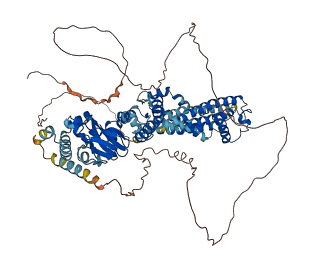
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
93-354 (Ion transport domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Target domain |
93-354 (Ion transport domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Wicks NL et al. (2011) "Cytoplasmic cAMP-sensing domain of hyperpolarization-activated cation (HCN) channels uses two structurally distinct mechanisms to regulate voltage gating", Proceedings of the National Academy of Sciences of the United States of America, 108, 609-14
- Akimoto M et al. (2014) "A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP", The Journal of biological chemistry, 289, 22205-20
- Xu X et al. (2010) "Structural basis for the cAMP-dependent gating in the human HCN4 channel", The Journal of biological chemistry, 285, 37082-91
Autoinhibited structure

Activated structure

1 structures for O88705
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O88705-F1 | Predicted | AlphaFoldDB |
33 variants for O88705
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388644665 | 110 | L>M | No | EVA | |
| rs3388629449 | 133 | D>G | No | EVA | |
| rs3388631523 | 186 | F>S | No | EVA | |
| rs228097029 | 251 | F>V | No | EVA | |
| rs3388645273 | 281 | C>R | No | EVA | |
| rs3388639283 | 290 | N>D | No | EVA | |
| rs3388635316 | 316 | Q>* | No | EVA | |
| rs3388643845 | 340 | M>V | No | EVA | |
| rs3388643857 | 377 | A>V | No | EVA | |
| rs3413089316 | 404 | E>G | No | EVA | |
| rs3388635776 | 404 | E>K | No | EVA | |
| rs3388635758 | 415 | N>T | No | EVA | |
| rs3388643880 | 437 | T>I | No | EVA | |
| rs3388645260 | 476 | A>T | No | EVA | |
| rs3388645256 | 498 | R>Q | No | EVA | |
| rs3388639230 | 519 | D>N | No | EVA | |
| rs3388629455 | 521 | F>S | No | EVA | |
| rs3388639274 | 527 | E>D | No | EVA | |
| rs3388634727 | 549 | K>R | No | EVA | |
| rs3388641726 | 567 | G>D | No | EVA | |
| rs3388639248 | 574 | V>L | No | EVA | |
| rs3388643840 | 575 | Q>* | No | EVA | |
| rs3388635736 | 576 | H>R | No | EVA | |
| rs3388635310 | 578 | R>I | No | EVA | |
| rs3388634726 | 579 | D>G | No | EVA | |
| rs3388639266 | 588 | A>P | No | EVA | |
| rs3388634783 | 645 | A>T | No | EVA | |
| rs3388642109 | 654 | S>F | No | EVA | |
| rs3388634786 | 656 | L>V | No | EVA | |
| rs3388642174 | 668 | G>E | No | EVA | |
| rs3388643884 | 673 | T>A | No | EVA | |
| rs3388643851 | 684 | L>I | No | EVA | |
| rs3388645244 | 717 | L>F | No | EVA |
No associated diseases with O88705
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cone cell pedicle | A specialized axon terminus which is produced by retinal cone cells. Pedicles are large, conical, flat end-feet (8-10 micrometers diameter) of the retinal cone axon that lie more or less side by side on the same plane at the outer edge of the outer plexiform layer (OPL). |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| HCN channel complex | A cation ion channel with a preference for K+ over Na+ ions, which is activated by membrane hyperpolarization, and consists of a tetramer of HCN family members. Some members of this family (HCN1, HCN2 and HCN4) are also activated when cAMP binds to their cyclic nucleotide binding domain (CNBD). Channel complexes of this family play an important role in the control of pacemaker activity in the heart. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| cAMP binding | Binding to cAMP, the nucleotide cyclic AMP (adenosine 3',5'-cyclophosphate). |
| voltage-gated potassium channel activity | Enables the transmembrane transfer of a potassium ion by a voltage-gated channel. A voltage-gated channel is a channel whose open state is dependent on the voltage across the membrane in which it is embedded. |
| voltage-gated sodium channel activity | Enables the transmembrane transfer of a sodium ion by a voltage-gated channel. A voltage-gated channel is a channel whose open state is dependent on the voltage across the membrane in which it is embedded. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to dopamine | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a dopamine stimulus. |
| potassium ion transmembrane transport | A process in which a potassium ion is transported from one side of a membrane to the other. |
| regulation of membrane depolarization | Any process that modulates the rate, frequency or extent of membrane depolarization. Membrane depolarization is the process in which membrane potential changes in the depolarizing direction from the resting potential, usually from negative to positive. |
| sodium ion transmembrane transport | A process in which a sodium ion is transported from one side of a membrane to the other by means of some agent such as a transporter or pore. |
18 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9UL51 | HCN2 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | Homo sapiens (Human) | SS |
| Q9Y3Q4 | HCN4 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | Homo sapiens (Human) | EV |
| O60741 | HCN1 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | Homo sapiens (Human) | SS |
| Q9P1Z3 | HCN3 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | Homo sapiens (Human) | SS |
| O88703 | Hcn2 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | Mus musculus (Mouse) | EV |
| O88704 | Hcn1 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | Mus musculus (Mouse) | SS |
| O70507 | Hcn4 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | Mus musculus (Mouse) | SS |
| Q9JKA9 | Hcn2 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | Rattus norvegicus (Rat) | SS |
| Q9JKA7 | Hcn4 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | Rattus norvegicus (Rat) | SS |
| Q9JKB0 | Hcn1 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | Rattus norvegicus (Rat) | SS |
| Q9JKA8 | Hcn3 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | Rattus norvegicus (Rat) | SS |
| A2ZX97 | Os01g0718700 | Potassium channel KAT6 | Oryza sativa subsp japonica (Rice) | PR |
| Q6K3T2 | Os02g0245800 | Potassium channel KAT1 | Oryza sativa subsp japonica (Rice) | PR |
| Q9SKD7 | CNGC3 | Probable cyclic nucleotide-gated ion channel 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LDR2 | CNGC19 | Putative cyclic nucleotide-gated ion channel 19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SU64 | CNGC16 | Probable cyclic nucleotide-gated ion channel 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P92960 | ATHB-4 | Potassium channel KAT3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39128 | KAT1 | Potassium channel KAT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEEEARPAAG | AGEAATPARE | TPPAAPAQAR | AASGGVPESA | PEPKRRQLGT | LLQPTVNKFS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LRVFGSHKAV | EIEQERVKSA | GAWIIHPYSD | FRFYWDLIML | LLMVGNLIVL | PVGITFFKEE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NSPPWIVFNV | LSDTFFLLDL | VLNFRTGIVV | EEGAEILLAP | RAIRTRYLRT | WFLVDLISSI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PVDYIFLVVE | LEPRLDAEVY | KTARALRIVR | FTKILSLLRL | LRLSRLIRYI | HQWEEIFHMT |
| 250 | 260 | 270 | 280 | 290 | 300 |
| YDLASAVVRI | FNLIGMMLLL | CHWDGCLQFL | VPMLQDFPSD | CWVSMNRMVN | HSWGRQYSHA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LFKAMSHMLC | IGYGQQAPVG | MPDVWLTMLS | MIVGATCYAM | FIGHATALIQ | SLDSSRRQYQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EKYKQVEQYM | SFHKLPADTR | QRIHEYYEHR | YQGKMFDEES | ILGELSEPLR | EEIINFTCRG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LVAHMPLFAH | ADPSFVTAVL | TKLRFEVFQP | GDLVVREGSV | GRKMYFIQHG | LLSVLARGAR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DTRLTDGSYF | GEICLLTRGR | RTASVRADTY | CRLYSLSVDH | FNAVLEEFPM | MRRAFETVAM |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DRLRRIGKKN | SILQRKRSEP | SPGSSGGVME | QHLVQHDRDM | ARGVRGLAPG | TGARLSGKPV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LWEPLVHAPL | QAAAVTSNVA | IALTHQRGPL | PLSPDSPATL | LARSARRSAG | SPASPLVPVR |
| 670 | 680 | 690 | 700 | 710 | 720 |
| AGPLLARGPW | ASTSRLPAPP | ARTLHASLSR | TGRSQVSLLG | PPPGGGARRL | GPRGRPLSAS |
| 730 | 740 | 750 | 760 | 770 | |
| QPSLPQRATG | DGSPRRKGSG | SERLPPSGLL | AKPPGTVQPP | RSSVPEPVTP | RGPQISANM |