O80722
Gene name |
PME4 (ARATH18, VGDH1, At2g47030, F14M4.14) |
Protein name |
Pectinesterase 4 |
Names |
PE 4, Pectin methylesterase 18, AtPME18, Pectin methylesterase 4, AtPME4, VANGUARD1-like protein 1, VGD1-like protein 1 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT2G47030 |
EC number |
3.1.1.11: Carboxylic ester hydrolases |
Protein Class |
|
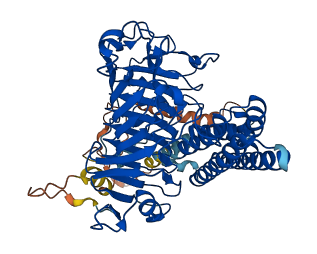
Descriptions
(Annotation from UniProt)
The PMEI region may act as an autoinhibitory domain.
Autoinhibitory domains (AIDs)
Target domain |
277-571 (Pectinesterase domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for O80722
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O80722-F1 | Predicted | AlphaFoldDB |
51 variants for O80722
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH05732776 | 5 | V>I | No | 1000Genomes | |
| ENSVATH13664563 | 9 | V>L | No | 1000Genomes | |
| ENSVATH05732773 | 20 | I>T | No | 1000Genomes | |
| tmp_2_19326180_C_T | 30 | G>D | No | 1000Genomes | |
| ENSVATH05732772 | 47 | S>A | No | 1000Genomes | |
| tmp_2_19326077_C_A | 64 | E>D | No | 1000Genomes | |
| ENSVATH14623448 | 71 | K>T | No | 1000Genomes | |
| tmp_2_19326047_C_T | 74 | M>I | No | 1000Genomes | |
| tmp_2_19326046_G_C | 75 | L>V | No | 1000Genomes | |
| tmp_2_19325991_A_C | 93 | V>G | No | 1000Genomes | |
| tmp_2_19325992_C_A | 93 | V>L | No | 1000Genomes | |
| tmp_2_19325928_A_G | 114 | F>S | No | 1000Genomes | |
| tmp_2_19325890_C_G | 127 | E>Q | No | 1000Genomes | |
| ENSVATH05732770 | 130 | E>D | No | 1000Genomes | |
| tmp_2_19325877_T_G | 131 | D>A | No | 1000Genomes | |
| ENSVATH05732769 | 135 | I>S | No | 1000Genomes | |
| tmp_2_19325860_T_A | 137 | S>C | No | 1000Genomes | |
| tmp_2_19325856_T_A | 138 | K>I | No | 1000Genomes | |
| tmp_2_19325845_G_A | 142 | L>F | No | 1000Genomes | |
| tmp_2_19325829_A_G | 147 | I>T | No | 1000Genomes | |
| ENSVATH14623447 | 185 | D>E | No | 1000Genomes | |
| ENSVATH13664562 | 195 | A>T | No | 1000Genomes | |
| ENSVATH14623445 | 198 | N>K | No | 1000Genomes | |
| ENSVATH14623444 | 200 | K>R | No | 1000Genomes | |
| ENSVATH13664561 | 217 | P>L | No | 1000Genomes | |
| ENSVATH01997215 | 219 | V>I | No | 1000Genomes | |
| tmp_2_19325493_G_A | 259 | A>V | No | 1000Genomes | |
| ENSVATH01997214 | 275 | T>A | No | 1000Genomes | |
| ENSVATH01997213 | 281 | K>E | No | 1000Genomes | |
| tmp_2_19325364_C_T | 302 | R>Q | No | 1000Genomes | |
| tmp_2_19325308_G_T | 321 | P>T | No | 1000Genomes | |
| ENSVATH01997212 | 324 | K>N | No | 1000Genomes | |
| tmp_2_19325294_GTTC_CTTC,G | 325 | N>K | No | 1000Genomes | |
| ENSVATH05732766 | 347 | L>I | No | 1000Genomes | |
| tmp_2_19325211_G_A | 353 | T>I | No | 1000Genomes | |
| tmp_2_19325194_C_T | 359 | V>I | No | 1000Genomes | |
| ENSVATH05732764 | 371 | M>I | No | 1000Genomes | |
| ENSVATH13664555 | 376 | T>I | No | 1000Genomes | |
| ENSVATH05732762 | 418 | R>G | No | 1000Genomes | |
| ENSVATH13664551 | 467 | K>* | No | 1000Genomes | |
| ENSVATH05732761 | 470 | I>V | No | 1000Genomes | |
| tmp_2_19324742_G_T | 480 | D>E | No | 1000Genomes | |
| ENSVATH14623443 | 503 | T>I | No | 1000Genomes | |
| ENSVATH13664550 | 507 | M>V | No | 1000Genomes | |
| tmp_2_19324516_C_A | 556 | A>S | No | 1000Genomes | |
| ENSVATH01997210 | 557 | R>K | No | 1000Genomes | |
| ENSVATH05732756 | 558 | S>T | No | 1000Genomes | |
| ENSVATH13664545 | 567 | A>V | No | 1000Genomes | |
| ENSVATH01997209 | 580 | A>T | No | 1000Genomes | |
| ENSVATH05732755 | 581 | N>H | No | 1000Genomes | |
| ENSVATH13664532 | 586 | I>L | No | 1000Genomes |
No associated diseases with O80722
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.1.11 | Carboxylic ester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| plasmodesma | A fine cytoplasmic channel, found in all higher plants, that connects the cytoplasm of one cell to that of an adjacent cell. |
| pollen tube | A tubular cell projection that is part of a pollen tube cell and extends from a pollen grain. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| aspartyl esterase activity | Catalysis of the hydrolysis of an ester bond by a mechanism involving a catalytically active aspartic acid residue. |
| pectinesterase activity | Catalysis of the reaction: pectin + n H2O = n methanol + pectate. |
| pectinesterase inhibitor activity | Binds to and stops, prevents or reduces the activity of pectinesterase. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell wall modification | The series of events leading to chemical and structural alterations of an existing cell wall that can result in loosening, increased extensibility or disassembly. |
| pectin catabolic process | The chemical reactions and pathways resulting in the breakdown of pectin, a polymer containing a backbone of alpha-1,4-linked D-galacturonic acid residues. |
42 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5MFV6 | VGDH2 | Probable pectinesterase/pectinesterase inhibitor VGDH2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV8 | PME5 | Pectinesterase 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SRX4 | PME7 | Probable pectinesterase/pectinesterase inhibitor 7 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1JPL7 | PME18 | Pectinesterase/pectinesterase inhibitor 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84JX1 | PME19 | Probable pectinesterase/pectinesterase inhibitor 19 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49298 | PME6 | Probable pectinesterase/pectinesterase inhibitor 6 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42534 | PME2 | Pectinesterase 2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q43867 | PME1 | Pectinesterase 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O48711 | PME12 | Probable pectinesterase/pectinesterase inhibitor 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q7Y201 | PME13 | Probable pectinesterase/pectinesterase inhibitor 13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SKX2 | PME16 | Probable pectinesterase/pectinesterase inhibitor 16 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22149 | PME17 | Probable pectinesterase/pectinesterase inhibitor 17 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22256 | PME20 | Probable pectinesterase/pectinesterase inhibitor 20 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GX86 | PME21 | Probable pectinesterase/pectinesterase inhibitor 21 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M9W7 | PME22 | Putative pectinesterase/pectinesterase inhibitor 22 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GXA1 | PME23 | Probable pectinesterase/pectinesterase inhibitor 23 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SG77 | PME24 | Putative pectinesterase/pectinesterase inhibitor 24 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49006 | PME3 | Pectinesterase/pectinesterase inhibitor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9STY3 | PME33 | Probable pectinesterase/pectinesterase inhibitor 33 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LYT5 | PME35 | Probable pectinesterase/pectinesterase inhibitor 35 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84R10 | PME36 | Probable pectinesterase/pectinesterase inhibitor 36 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81320 | PME38 | Putative pectinesterase/pectinesterase inhibitor 38 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81415 | PME39 | Probable pectinesterase/pectinesterase inhibitor 39 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81301 | PME40 | Probable pectinesterase/pectinesterase inhibitor 40 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8RXK7 | PME41 | Probable pectinesterase/pectinesterase inhibitor 41 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1PEC0 | PME42 | Probable pectinesterase/pectinesterase inhibitor 42 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O23447 | PME43 | Putative pectinesterase/pectinesterase inhibitor 43 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY7 | PME44 | Probable pectinesterase/pectinesterase inhibitor 44 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY6 | PME45 | Putative pectinesterase/pectinesterase inhibitor 45 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF78 | PME46 | Probable pectinesterase/pectinesterase inhibitor 46 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF77 | PME47 | Probable pectinesterase/pectinesterase inhibitor 47 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXD9 | PME51 | Probable pectinesterase/pectinesterase inhibitor 51 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E989 | PME54 | Probable pectinesterase/pectinesterase inhibitor 54 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E8Z8 | PME28 | Putative pectinesterase/pectinesterase inhibitor 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FJ21 | PME58 | Probable pectinesterase/pectinesterase inhibitor 58 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN5 | PME59 | Probable pectinesterase/pectinesterase inhibitor 59 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN4 | PME60 | Probable pectinesterase/pectinesterase inhibitor 60 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FK05 | PME61 | Probable pectinesterase/pectinesterase inhibitor 61 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8L7Q7 | PME64 | Probable pectinesterase/pectinesterase inhibitor 64 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q94CB1 | PME25 | Probable pectinesterase/pectinesterase inhibitor 25 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXK7 | PME32 | Probable pectinesterase/pectinesterase inhibitor 32 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M3B0 | PME34 | Probable pectinesterase/pectinesterase inhibitor 34 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MIGKVVVSVA | SILLIVGVAI | GVVAFINKNG | DANLSPQMKA | VQGICQSTSD | KASCVKTLEP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VKSEDPNKLI | KAFMLATKDE | LTKSSNFTGQ | TEVNMGSSIS | PNNKAVLDYC | KRVFMYALED |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LATIIEEMGE | DLSQIGSKID | QLKQWLIGVY | NYQTDCLDDI | EEDDLRKAIG | EGIANSKILT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TNAIDIFHTV | VSAMAKINNK | VDDLKNMTGG | IPTPGAPPVV | DESPVADPDG | PARRLLEDID |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ETGIPTWVSG | ADRKLMAKAG | RGRRGGRGGG | ARVRTNFVVA | KDGSGQFKTV | QQAVDACPEN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NRGRCIIYIK | AGLYREQVII | PKKKNNIFMF | GDGARKTVIS | YNRSVALSRG | TTTSLSATVQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VESEGFMAKW | MGFKNTAGPM | GHQAAAIRVN | GDRAVIFNCR | FDGYQDTLYV | NNGRQFYRNC |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VVSGTVDFIF | GKSATVIQNT | LIVVRKGSKG | QYNTVTADGN | ELGLGMKIGI | VLQNCRIVPD |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RKLTPERLTV | ATYLGRPWKK | FSTTVIMSTE | MGDLIRPEGW | KIWDGESFHK | SCRYVEYNNR |
| 550 | 560 | 570 | 580 | ||
| GPGAFANRRV | NWAKVARSAA | EVNGFTAANW | LGPINWIQEA | NVPVTIGL |