O75909
Gene name |
CCNK (CPR4) |
Protein name |
Cyclin-K |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:8812 |
EC number |
|
Protein Class |
|
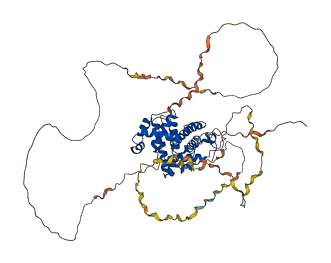
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

41 structures for O75909
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2I53 | X-ray | 150 A | A | 11-267 | PDB |
| 4CXA | X-ray | 315 A | B/D | 11-267 | PDB |
| 4NST | X-ray | 220 A | B/D | 1-267 | PDB |
| 4UN0 | X-ray | 315 A | A/B | 11-267 | PDB |
| 5ACB | X-ray | 270 A | A/B | 11-267 | PDB |
| 5EFQ | X-ray | 200 A | B/D | 1-267 | PDB |
| 6B3E | X-ray | 306 A | B/D | 1-267 | PDB |
| 6CKX | X-ray | 280 A | B/D | 1-267 | PDB |
| 6TD3 | X-ray | 346 A | C/F/I | 1-267 | PDB |
| 7NXJ | X-ray | 236 A | B/D | 1-267 | PDB |
| 7NXK | X-ray | 300 A | B/D | 1-267 | PDB |
| 8BU1 | X-ray | 298 A | C/F/I | 1-267 | PDB |
| 8BU2 | X-ray | 313 A | C/F/I | 1-267 | PDB |
| 8BU3 | X-ray | 342 A | C/F/I | 1-267 | PDB |
| 8BU4 | X-ray | 309 A | C/F/I | 1-267 | PDB |
| 8BU5 | X-ray | 313 A | C/F/I | 1-267 | PDB |
| 8BU6 | X-ray | 345 A | C/F/I | 1-267 | PDB |
| 8BU7 | X-ray | 325 A | C/F/I | 1-267 | PDB |
| 8BU9 | X-ray | 351 A | C/F/I | 1-267 | PDB |
| 8BUA | X-ray | 319 A | C/F/I | 1-267 | PDB |
| 8BUB | X-ray | 342 A | C/F/I | 1-267 | PDB |
| 8BUC | X-ray | 385 A | C/F/I | 1-267 | PDB |
| 8BUD | X-ray | 320 A | C/F/I | 1-267 | PDB |
| 8BUE | X-ray | 325 A | C/F/I | 1-267 | PDB |
| 8BUF | X-ray | 330 A | C/F/I | 1-267 | PDB |
| 8BUG | X-ray | 353 A | C/F/I | 1-267 | PDB |
| 8BUH | X-ray | 379 A | C/F/I | 1-267 | PDB |
| 8BUI | X-ray | 350 A | C/F/I | 1-267 | PDB |
| 8BUJ | X-ray | 362 A | C/F/I | 1-267 | PDB |
| 8BUK | X-ray | 341 A | C/F/I | 1-267 | PDB |
| 8BUL | X-ray | 340 A | C/F/I | 1-267 | PDB |
| 8BUM | X-ray | 336 A | C/F/I | 1-267 | PDB |
| 8BUN | X-ray | 308 A | C/F/I | 1-267 | PDB |
| 8BUO | X-ray | 358 A | C/F/I | 1-267 | PDB |
| 8BUP | X-ray | 341 A | C/F/I | 1-267 | PDB |
| 8BUQ | X-ray | 320 A | C/F/I | 1-267 | PDB |
| 8BUR | X-ray | 364 A | C/F/I | 1-267 | PDB |
| 8BUS | X-ray | 326 A | C/F/I | 1-267 | PDB |
| 8BUT | X-ray | 325 A | C/F/I | 1-267 | PDB |
| 8P81 | X-ray | 268 A | B | 1-267 | PDB |
| AF-O75909-F1 | Predicted | AlphaFoldDB |
327 variants for O75909
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
VAR_081570 RCV000710289 rs1566748800 CA390942452 |
111 | K>E | Intellectual developmental disorder with hypertelorism and distinctive facies IDDHDF; contrary to the wild-type protein, does not rescue morpholino knockdown phenotype in zebrafish [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001291721 CA7340685 rs750789680 |
246 | V>I | Intellectual developmental disorder with hypertelorism and distinctive facies [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA390941362 rs1340506299 |
9 | S>G | No |
ClinGen TOPMed |
|
|
rs1475416350 CA390941366 |
9 | S>T | No |
ClinGen gnomAD |
|
|
rs890479145 CA266133239 |
10 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1002256892 CA266133242 |
12 | V>G | No |
ClinGen Ensembl |
|
|
rs1595307990 CA390941390 |
12 | V>I | No |
ClinGen Ensembl |
|
| TCGA novel | 13 | T>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs538109875 CA7340549 |
15 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA390941434 rs1304131376 |
16 | N>D | No |
ClinGen gnomAD |
|
|
CA7340550 rs781186631 |
17 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs1203186888 CA390941528 |
22 | P>L | No |
ClinGen gnomAD |
|
|
rs749217487 CA7340551 |
23 | C>G | No |
ClinGen ExAC gnomAD |
|
|
rs1245373903 CA390941560 |
25 | Y>C | No |
ClinGen gnomAD |
|
|
rs1366612744 CA390941565 |
26 | W>G | No |
ClinGen TOPMed |
|
|
CA390941617 rs1413745510 |
32 | A>V | No |
ClinGen gnomAD |
|
|
rs1173996260 CA390941622 |
33 | H>L | No |
ClinGen gnomAD |
|
| TCGA novel | 33 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 35 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 35 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs778610231 CA7340553 |
39 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1455191065 CA390941665 |
40 | G>R | No |
ClinGen gnomAD |
|
|
CA266133250 rs964869039 |
46 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs958424865 CA266133254 |
48 | R>Q | No |
ClinGen Ensembl |
|
|
rs1377006286 CA390941716 |
48 | R>W | No |
ClinGen gnomAD |
|
|
CA390941730 rs1326488496 |
50 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1226198377 CA390941736 |
51 | R>Q | No |
ClinGen gnomAD |
|
|
rs1269695112 CA390941739 |
52 | E>K | No |
ClinGen gnomAD |
|
|
rs749729510 CA266133257 |
54 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1288054162 CA390941762 |
55 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1255369135 CA390941815 |
63 | R>H | No |
ClinGen gnomAD |
|
| TCGA novel | 66 | L>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA266133263 rs200721397 |
66 | L>Q | No |
ClinGen 1000Genomes |
|
| TCGA novel | 69 | D>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1419720129 CA390941877 |
71 | L>M | No |
ClinGen gnomAD |
|
|
rs868803509 CA266133378 |
72 | A>T | No |
ClinGen Ensembl |
|
|
CA390941891 rs1249577631 |
73 | T>S | No |
ClinGen Ensembl |
|
|
CA7340582 rs552001718 |
76 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1330833967 CA390941938 |
80 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs774653559 CA7340583 |
81 | F>V | No |
ClinGen ExAC gnomAD |
|
|
CA390941960 rs1402453647 |
83 | M>R | No |
ClinGen gnomAD |
|
|
rs767771543 CA7340585 |
88 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1479461683 CA390942283 |
98 | C>Y | No |
ClinGen gnomAD |
|
|
CA266133756 rs868692667 |
105 | K>E | No |
ClinGen Ensembl |
|
|
rs1158034053 CA390942444 |
110 | P>Q | No |
ClinGen gnomAD |
|
|
CA390942537 rs1191649509 |
118 | K>R | No |
ClinGen gnomAD |
|
|
CA390942542 rs1214276402 |
119 | T>A | No |
ClinGen TOPMed |
|
|
rs935043382 CA266133758 |
121 | R>C | No |
ClinGen Ensembl |
|
|
CA7340614 rs190257611 |
121 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA390942591 rs1417190597 |
125 | N>S | No |
ClinGen Ensembl |
|
|
CA7340617 rs368952076 |
126 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1394333927 CA390942615 |
127 | V>I | No |
ClinGen TOPMed |
|
|
rs1470435323 CA390942922 |
151 | I>V | No |
ClinGen TOPMed |
|
|
rs868057477 CA266134552 |
154 | D>Y | No |
ClinGen Ensembl |
|
|
CA266134553 rs868868559 |
160 | P>Q | No |
ClinGen Ensembl |
|
|
rs1443796779 CA390943034 |
166 | K>N | No |
ClinGen gnomAD |
|
|
CA7340633 rs767370737 |
167 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs746022344 CA7340647 |
177 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs982842489 CA266134616 |
191 | D>N | No |
ClinGen TOPMed |
|
|
rs1333281843 CA390943298 |
200 | Q>R | No |
ClinGen gnomAD |
|
|
CA390943308 rs1368710353 |
201 | W>* | No |
ClinGen TOPMed gnomAD |
|
|
CA390943312 rs1294516729 |
202 | E>* | No |
ClinGen TOPMed |
|
| TCGA novel | 206 | I>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA390943377 rs1475602740 |
211 | M>I | No |
ClinGen gnomAD |
|
|
CA266134694 rs962279113 |
212 | Y>D | No |
ClinGen Ensembl |
|
|
CA7340674 rs760617667 |
214 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7340675 rs370608733 |
216 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA390943429 rs1200919119 |
219 | K>I | No |
ClinGen gnomAD |
|
|
CA390943439 rs1468444980 |
220 | F>L | No |
ClinGen gnomAD |
|
|
rs1251614558 CA390943436 |
220 | F>S | No |
ClinGen gnomAD |
|
|
CA7340676 rs776386794 |
228 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA7340677 rs572139668 |
230 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390943517 rs1165780200 |
231 | Y>C | No |
ClinGen gnomAD |
|
|
CA390943584 rs1267750574 |
240 | Q>E | No |
ClinGen TOPMed |
|
|
CA7340680 rs762383866 |
243 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs767256145 CA7340683 |
245 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs767256145 CA390943616 |
245 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA390943632 rs1277670436 |
247 | L>S | No |
ClinGen TOPMed |
|
|
CA390943829 rs1325330474 |
267 | H>Y | No |
ClinGen TOPMed |
|
|
CA266134730 rs372216197 |
269 | T>N | No |
ClinGen ESP |
|
|
CA390943845 rs1595317080 |
269 | T>P | No |
ClinGen Ensembl |
|
|
CA390943856 rs1385735105 |
271 | H>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1375078716 CA390943909 |
278 | S>C | No |
ClinGen gnomAD |
|
|
rs1375078716 CA390943908 |
278 | S>Y | No |
ClinGen gnomAD |
|
|
CA7340715 rs748943823 |
283 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs768325984 CA7340716 |
284 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs774074339 CA7340717 |
285 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA7340718 rs200034824 |
286 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 286 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA266134732 rs372907456 |
289 | Q>* | No |
ClinGen ESP |
|
|
CA7340720 rs773882638 |
290 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs376026947 CA7340721 |
293 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs935270794 CA266134733 |
294 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
rs754196878 CA7340723 |
297 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1380915763 CA390944032 |
298 | E>K | No |
ClinGen gnomAD |
|
|
rs1319314451 CA390944041 |
299 | P>A | No |
ClinGen TOPMed |
|
|
CA390944048 rs1595317264 |
300 | S>P | No |
ClinGen Ensembl |
|
|
CA7340726 rs752858323 |
302 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA266134734 rs931964755 |
302 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA390944093 rs1595317306 |
306 | D>A | No |
ClinGen Ensembl |
|
|
CA390944094 rs1172393068 |
306 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs758477791 CA7340727 |
307 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1595317346 CA390944115 |
309 | Q>H | No |
ClinGen Ensembl |
|
|
rs534989573 CA7340731 |
312 | Q>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA390944143 rs1376395802 |
313 | Q>H | No |
ClinGen gnomAD |
|
|
CA390944150 rs1379776671 |
314 | Q>H | No |
ClinGen gnomAD |
|
|
CA390944148 rs1330059608 |
314 | Q>P | No |
ClinGen gnomAD |
|
|
rs1330308822 CA390944161 |
316 | P>A | No |
ClinGen TOPMed |
|
|
CA7340733 rs200877327 |
318 | Q>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs749107194 CA7340734 |
318 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs1460094258 CA390944180 |
319 | Q>E | No |
ClinGen TOPMed |
|
|
CA390944181 rs1296820172 |
319 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1346574897 CA390944193 |
321 | K>E | No |
ClinGen gnomAD |
|
|
rs1595317442 CA390944206 |
322 | K>N | No |
ClinGen Ensembl |
|
|
rs751012933 CA7340737 |
325 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778725600 CA7340736 |
325 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs771726550 CA7340738 |
331 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs761124604 CA7340740 |
332 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1191788942 CA390944265 |
332 | Q>K | No |
ClinGen gnomAD |
|
|
CA7340741 rs771462544 |
333 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA266134735 rs200503921 |
334 | K>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA7340742 rs200503921 |
334 | K>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs759957278 CA7340743 |
335 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs752948207 CA7340745 |
337 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752948207 CA266134736 |
337 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767434647 CA390944407 |
338 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs767434647 CA7340768 |
338 | V>D | No |
ClinGen ExAC gnomAD |
|
|
CA266134824 rs867171281 |
339 | V>F | No |
ClinGen gnomAD |
|
|
CA390944410 rs867171281 |
339 | V>I | No |
ClinGen gnomAD |
|
| TCGA novel | 340 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs868099511 CA266134825 |
343 | E>* | No |
ClinGen Ensembl |
|
|
CA266134826 rs934877498 |
343 | E>G | No |
ClinGen gnomAD |
|
|
CA390944500 rs1384731263 |
346 | K>Q | No |
ClinGen gnomAD |
|
|
rs753831877 CA7340769 |
346 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA266134828 rs1052124570 |
347 | A>V | No |
ClinGen Ensembl |
|
|
rs867095987 CA266134829 |
348 | A>E | No |
ClinGen Ensembl |
|
|
CA266134830 rs867531582 |
349 | E>* | No |
ClinGen Ensembl |
|
|
CA390944567 rs1324333622 |
350 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA7340784 rs775111606 |
351 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390944573 rs1307601966 |
351 | P>S | No |
ClinGen TOPMed |
|
|
CA7340785 rs749646615 |
352 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1428836044 CA390944578 |
352 | P>S | No |
ClinGen gnomAD |
|
|
CA390944586 rs1212784508 |
353 | P>L | No |
ClinGen TOPMed |
|
|
rs1220922392 CA390944619 |
358 | I>T | No |
ClinGen gnomAD |
|
|
CA7340786 rs768938736 |
358 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774643695 CA7340787 |
360 | T>N | No |
ClinGen ExAC TOPMed |
|
|
CA7340789 COSM959484 rs532436153 |
361 | T>A | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA390944637 rs901901957 |
361 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA7340790 rs532436153 |
361 | T>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA266135209 rs901901957 |
361 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1301552792 CA390944640 |
362 | H>Y | No |
ClinGen TOPMed |
|
|
CA390944646 rs1289919737 |
363 | P>S | No |
ClinGen gnomAD |
|
|
CA7340791 COSM959485 rs759607767 |
364 | P>L | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs752617499 CA7340793 |
368 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1595330668 CA390944722 |
373 | D>A | No |
ClinGen Ensembl |
|
|
rs779486624 CA266135578 |
374 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779486624 CA7340819 |
374 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7340818 rs755631919 |
374 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs199586655 CA266135579 |
375 | K>R | No |
ClinGen 1000Genomes |
|
|
rs1052057347 CA266135581 |
376 | P>L | No |
ClinGen Ensembl |
|
|
CA266135580 rs1052057347 |
376 | P>R | No |
ClinGen Ensembl |
|
|
rs1156717530 CA390944747 |
378 | L>V | No |
ClinGen TOPMed |
|
|
rs1258553086 CA390944756 |
379 | A>D | No |
ClinGen TOPMed |
|
|
CA266135582 rs912563511 |
379 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs754369845 CA7340820 |
380 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA390944772 rs1307853777 |
382 | L>S | No |
ClinGen gnomAD |
|
|
rs1353830668 CA390944775 |
383 | G>S | No |
ClinGen gnomAD |
|
|
rs1241798953 CA390944789 |
385 | A>T | No |
ClinGen gnomAD |
|
|
rs755459069 CA7340821 |
387 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390944805 rs1344031784 |
387 | P>S | No |
ClinGen gnomAD |
|
|
CA390944810 rs1485386934 |
388 | P>A | No |
ClinGen TOPMed |
|
|
rs779396416 CA7340822 |
388 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390944816 rs1238813636 |
389 | G>D | No |
ClinGen gnomAD |
|
|
rs1441637129 CA390944820 |
390 | P>A | No |
ClinGen gnomAD |
|
|
CA390944828 rs1196940660 |
391 | V>E | No |
ClinGen gnomAD |
|
|
CA266135585 rs899052490 |
392 | D>N | No |
ClinGen Ensembl |
|
|
rs1595330814 CA390944852 |
395 | D>A | No |
ClinGen Ensembl |
|
|
CA266135586 rs993472154 |
395 | D>N | No |
ClinGen Ensembl |
|
|
rs772417001 CA7340824 |
396 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA390944869 rs1174724135 |
398 | K>E | No |
ClinGen gnomAD |
|
|
rs1332648804 CA390944897 |
402 | P>T | No |
ClinGen gnomAD |
|
|
rs747181950 CA390944908 |
403 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA7340826 rs747181950 |
403 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA7340830 rs771166710 |
405 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA7340833 rs867004709 |
406 | H>P | No |
ClinGen TOPMed gnomAD |
|
|
CA7340837 rs369022783 |
407 | P>L | No |
ClinGen ESP ExAC TOPMed |
|
|
CA7340838 rs369022783 |
407 | P>Q | No |
ClinGen ESP ExAC TOPMed |
|
|
rs1400046688 CA390944975 |
408 | A>G | No |
ClinGen TOPMed |
|
|
CA390944977 rs1400046688 |
408 | A>V | No |
ClinGen TOPMed |
|
|
rs761515450 CA7340843 |
409 | P>L | No |
ClinGen ExAC |
|
| TCGA novel | 410 | V>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA390944995 CA390944993 rs1315861367 |
410 | V>L | No |
ClinGen gnomAD |
|
|
rs900838475 CA266135589 |
411 | H>P | No |
ClinGen TOPMed gnomAD |
|
|
rs900838475 CA390945013 |
411 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1459139101 CA390945026 |
412 | Q>L | No |
ClinGen gnomAD |
|
|
CA7340844 rs767282814 |
413 | P>Q | No |
ClinGen ExAC |
|
|
rs750096195 CA7340845 |
414 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7340847 rs535214522 |
415 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed |
|
|
rs779295552 CA390945092 |
419 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7340850 rs779295552 |
419 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7340849 rs376159668 |
419 | R>W | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1595331051 CA390945095 |
420 | P>A | No |
ClinGen Ensembl |
|
|
rs758713375 CA7340852 |
421 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA390945103 rs1376841644 |
421 | P>L | No |
ClinGen gnomAD |
|
|
CA390945104 rs1376841644 |
421 | P>R | No |
ClinGen gnomAD |
|
|
CA7340855 rs757529597 |
422 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390945107 rs1403160623 |
422 | P>S | No |
ClinGen gnomAD |
|
|
rs781478973 CA7340856 |
423 | P>L | No |
ClinGen ExAC |
|
|
rs781478973 CA390945114 |
423 | P>R | No |
ClinGen ExAC |
|
|
rs1233031038 CA390945110 |
423 | P>T | No |
ClinGen TOPMed |
|
|
rs1218732319 CA390945120 |
424 | P>L | No |
ClinGen TOPMed |
|
|
rs1279653590 CA390945115 |
424 | P>T | No |
ClinGen TOPMed |
|
|
CA7340858 rs769843517 |
425 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769843517 CA7340859 |
425 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390945121 rs769843517 |
425 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7340860 rs748067534 |
426 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA7340861 rs771790935 |
426 | S>Y | No |
ClinGen ExAC |
|
|
CA7340862 rs772955703 |
427 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA7340863 rs760381998 |
427 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs1217851623 CA390945136 |
428 | Y>H | No |
ClinGen gnomAD |
|
|
CA266135591 rs1003543184 |
429 | M>K | No |
ClinGen TOPMed |
|
|
CA7340864 rs766019646 |
429 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1242655843 CA390945170 |
430 | T>S | No |
ClinGen gnomAD |
|
|
CA7340866 rs758984649 |
431 | G>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA390945207 rs1261390147 |
433 | S>A | No |
ClinGen gnomAD |
|
|
CA390945215 rs1488160670 |
434 | T>A | No |
ClinGen gnomAD |
|
|
rs1488160670 CA390945214 |
434 | T>P | No |
ClinGen gnomAD |
|
|
rs1194908550 CA390945221 |
434 | T>S | No |
ClinGen gnomAD |
|
|
CA390945284 rs1367318907 |
439 | M>I | No |
ClinGen gnomAD |
|
|
CA7340868 rs753176466 |
439 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA390945288 rs1173027334 |
440 | S>T | No |
ClinGen TOPMed |
|
|
CA390945298 rs1304576863 |
441 | G>R | No |
ClinGen gnomAD |
|
|
rs1399137326 CA390945316 |
442 | E>G | No |
ClinGen gnomAD |
|
| TCGA novel | 444 | Y>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7340869 rs201501099 |
445 | Q>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1363849142 CA390945371 |
446 | S>R | No |
ClinGen gnomAD |
|
|
CA390945423 rs1322405212 |
450 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1311933968 CA390945415 |
450 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
CA390945414 rs1311933968 |
450 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1247158712 TCGA novel CA390945442 |
451 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen gnomAD NCI-TCGA |
| TCGA novel | 451 | M>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs894145157 CA266135594 |
452 | K>Q | No |
ClinGen TOPMed |
|
|
rs867757117 CA266135595 |
454 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1217663367 CA390945484 |
455 | G>R | No |
ClinGen gnomAD |
|
|
CA266135596 rs970250072 |
456 | P>A | No |
ClinGen gnomAD |
|
|
rs1595331377 CA390945505 |
457 | S>A | No |
ClinGen Ensembl |
|
| TCGA novel | 458 | Y>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7340873 rs565765320 |
459 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1595331390 CA390945534 |
459 | G>V | No |
ClinGen Ensembl |
|
|
rs1198806194 CA390945545 |
460 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs963110983 CA266135598 |
462 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA390945569 rs1331644229 |
463 | P>A | No |
ClinGen TOPMed |
|
|
rs746043550 CA7340874 |
463 | P>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 468 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA266135599 rs997139602 |
473 | Y>S | No |
ClinGen Ensembl |
|
|
rs1028260055 CA266135600 |
474 | H>P | No |
ClinGen TOPMed gnomAD |
|
|
CA266135601 rs952529266 |
476 | H>P | No |
ClinGen Ensembl |
|
|
CA390945750 rs1278356047 |
478 | Y>* | No |
ClinGen Ensembl |
|
|
CA390945746 rs534601080 |
478 | Y>C | No |
ClinGen 1000Genomes |
|
|
CA266135602 rs534601080 |
478 | Y>S | No |
ClinGen 1000Genomes |
|
|
rs1015626086 CA266135603 |
481 | N>T | No |
ClinGen TOPMed |
|
|
CA390945791 rs1376284414 |
482 | P>L | No |
ClinGen gnomAD |
|
|
rs1288430029 CA390945813 |
484 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1595331533 CA390945866 |
489 | P>R | No |
ClinGen Ensembl |
|
|
CA390945873 rs1257072835 |
490 | P>S | No |
ClinGen gnomAD |
|
|
rs1177345987 CA390945882 |
491 | P>S | No |
ClinGen gnomAD |
|
|
CA390945902 rs1595331574 |
493 | S>P | No |
ClinGen Ensembl |
|
|
CA390945910 rs1595331581 |
494 | F>L | No |
ClinGen Ensembl |
|
|
CA390945915 rs1232085524 |
494 | F>S | No |
ClinGen TOPMed |
|
|
rs1412522062 CA390945935 |
496 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1412522062 CA390945932 |
496 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1412522062 CA390945934 |
496 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs866279123 CA266135604 |
499 | I>T | No |
ClinGen Ensembl |
|
|
rs112286214 CA266135605 |
500 | P>R | No |
ClinGen Ensembl |
|
|
rs1376073104 CA390946004 |
503 | T>A | No |
ClinGen TOPMed |
|
|
CA390946010 rs1470888312 |
503 | T>I | No |
ClinGen gnomAD |
|
|
rs1376073104 CA390946002 |
503 | T>P | No |
ClinGen TOPMed |
|
|
rs1173084351 CA390946037 |
506 | Y>S | No |
ClinGen TOPMed |
|
|
CA390946048 rs1329242584 |
507 | P>R | No |
ClinGen gnomAD |
|
|
CA390946088 rs764694208 |
512 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1595331696 CA390946092 |
512 | T>I | No |
ClinGen Ensembl |
|
|
CA7340884 rs764694208 |
512 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390946118 rs1230834346 |
514 | N>K | No |
ClinGen TOPMed |
|
|
CA390946114 rs1469905084 |
514 | N>S | No |
ClinGen TOPMed |
|
|
CA390946112 rs1469905084 |
514 | N>T | No |
ClinGen TOPMed |
|
|
rs866744998 CA266135606 |
516 | N>H | No |
ClinGen Ensembl |
|
|
CA390946139 rs1595331736 |
516 | N>K | No |
ClinGen Ensembl |
|
|
rs1595331728 CA390946133 |
516 | N>T | No |
ClinGen Ensembl |
|
|
rs1189112911 CA390946142 |
517 | F>I | No |
ClinGen TOPMed |
|
|
CA390946143 rs1189112911 |
517 | F>L | No |
ClinGen TOPMed |
|
|
CA266135607 rs868786858 |
517 | F>S | No |
ClinGen Ensembl |
|
|
rs868786858 CA390946147 |
517 | F>Y | No |
ClinGen Ensembl |
|
| TCGA novel | 520 | P>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1456922960 CA390946205 |
523 | R>H | No |
ClinGen gnomAD |
|
|
rs1456922960 CA390946207 |
523 | R>P | No |
ClinGen gnomAD |
|
|
rs1198784842 CA390946217 |
524 | L>P | No |
ClinGen gnomAD |
|
|
rs954617656 CA390946258 |
528 | H>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA390946260 rs1425537573 |
529 | A>T | No |
ClinGen gnomAD |
|
|
CA390946302 rs1422901475 |
533 | H>P | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 536 | P>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1212307981 CA390946332 |
536 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA390946359 rs1319492553 |
539 | G>C | No |
ClinGen TOPMed |
|
|
CA390946379 rs1466186352 |
541 | P>L | No |
ClinGen gnomAD |
|
|
CA390946381 rs1406326892 |
542 | P>T | No |
ClinGen TOPMed |
|
|
rs1435962560 CA390946395 |
543 | A>D | No |
ClinGen gnomAD |
|
|
CA7340889 rs752043596 |
544 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs1475361156 CA390946433 |
547 | P>S | No |
ClinGen TOPMed |
|
|
rs577528220 CA7340892 |
550 | V>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA7340891 rs374537883 |
550 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1242643284 CA390946466 |
551 | P>T | No |
ClinGen TOPMed |
|
|
CA390946477 rs753951143 |
552 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7340895 rs753951143 |
552 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA390946548 rs1433868267 |
559 | P>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1595331914 CA390946554 |
560 | P>A | No |
ClinGen Ensembl |
|
|
CA7340899 rs770745891 |
560 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA7340901 rs745557007 |
561 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA266135612 rs868509054 |
566 | G>D | No |
ClinGen gnomAD |
|
|
rs202078095 CA7340904 |
566 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA390946651 rs1398513000 |
570 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
rs368703834 CA7340906 |
571 | G>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7340907 rs368703834 |
571 | G>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA390946657 rs1430718080 |
571 | G>W | No |
ClinGen TOPMed |
|
|
rs762366193 CA7340908 |
575 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs762366193 CA7340909 |
575 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs750774367 CA7340910 |
576 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1041854216 CA266135613 |
577 | A>T | No |
ClinGen TOPMed |
|
| TCGA novel | 579 | M>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
1 associated diseases with O75909
[MIM: 618147]: Intellectual developmental disorder with hypertelorism and distinctive facies (IDDHDF)
An autosomal dominant neurodevelopmental disorder characterized by developmental delay and intellectual disability, language defects, and distinctive facial dysmorphism including high hairline, hypertelorism, thin eyebrows, dysmorphic ears, broad nasal bridge and tip, and narrow jaw. {ECO:0000269|PubMed:30122539}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal dominant neurodevelopmental disorder characterized by developmental delay and intellectual disability, language defects, and distinctive facial dysmorphism including high hairline, hypertelorism, thin eyebrows, dysmorphic ears, broad nasal bridge and tip, and narrow jaw. {ECO:0000269|PubMed:30122539}. Note=The disease is caused by variants affecting the gene represented in this entry.
4 regional properties for O75909
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Cyclin, C-terminal domain | 158 - 278 | IPR004367 |
| domain | Cyclin, N-terminal | 42 - 155 | IPR006671 |
| domain | Cyclin-like domain | 55 - 149 | IPR013763-1 |
| domain | Cyclin-like domain | 162 - 256 | IPR013763-2 |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cyclin K-CDK12 complex | A protein complex consisting of cyclin Kand cyclin-dependent kinase 12 (CDK12). Cyclins are characterized by periodicity in protein abundance throughout the cell cycle. Cyclin-dependent kinases represent a family of serine/threonine protein kinases that become active upon binding to a cyclin regulatory partner. |
| cyclin K-CDK13 complex | A protein complex consisting of cyclin Kand cyclin-dependent kinase 13 (CDK13). Cyclins are characterized by periodicity in protein abundance throughout the cell cycle. Cyclin-dependent kinases represent a family of serine/threonine protein kinases that become active upon binding to a cyclin regulatory partner. |
| cyclin/CDK positive transcription elongation factor complex | A transcription elongation factor complex that facilitates the transition from abortive to productive elongation by phosphorylating the CTD domain of the large subunit of DNA-directed RNA polymerase II, holoenzyme. Contains a cyclin and a cyclin-dependent protein kinase catalytic subunit. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| cyclin-dependent protein serine/threonine kinase activator activity | Binds to and increases the activity of a cyclin-dependent protein serine/threonine kinase. |
| cyclin-dependent protein serine/threonine kinase activity | Cyclin-dependent catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| cyclin-dependent protein serine/threonine kinase regulator activity | Modulates the activity of a cyclin-dependent protein serine/threonine kinase, enzymes of the protein kinase family that are regulated through association with cyclins and other proteins. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| RNA polymerase II CTD heptapeptide repeat kinase activity | Catalysis of the reaction: ATP + RNA polymerase II large subunit CTD heptapeptide repeat (YSPTSPS) = ADP + H+ + phosphorylated RNA polymerase II. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| cellular response to DNA damage stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
| negative regulation by host of viral genome replication | A process in which a host organism stops, prevents or reduces the frequency, rate or extent of viral genome replication. |
| phosphorylation of RNA polymerase II C-terminal domain | The process of introducing a phosphate group on to an amino acid residue in the C-terminal domain of RNA polymerase II. Typically, this occurs during the transcription cycle and results in production of an RNA polymerase II enzyme where the carboxy-terminal domain (CTD) of the largest subunit is extensively phosphorylated, often referred to as hyperphosphorylated or the II(0) form. Specific types of phosphorylation within the CTD are usually associated with specific regions of genes, though there are exceptions. The phosphorylation state regulates the association of specific complexes such as the capping enzyme or 3'-RNA processing machinery to the elongating RNA polymerase complex. |
| positive regulation of DNA-templated transcription, elongation | Any process that activates or increases the frequency, rate or extent of transcription elongation, the extension of an RNA molecule after transcription initiation and promoter clearance by the addition of ribonucleotides catalyzed by a DNA-dependent RNA polymerase. |
| positive regulation of phosphorylation of RNA polymerase II C-terminal domain serine 2 residues | Any process that activates or increases the frequency, rate or extent of phosphorylation of RNA polymerase II C-terminal domain serine 2 residues. |
| positive regulation of transcription elongation by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription elongation, the extension of an RNA molecule after transcription initiation and promoter clearance by the addition of ribonucleotides, catalyzed by RNA polymerase II. |
| regulation of cyclin-dependent protein serine/threonine kinase activity | Any process that modulates the frequency, rate or extent of cyclin-dependent protein serine/threonine kinase activity. |
| regulation of signal transduction | Any process that modulates the frequency, rate or extent of signal transduction. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| transcription by RNA polymerase II | The synthesis of RNA from a DNA template by RNA polymerase II (RNAP II), originating at an RNA polymerase II promoter. Includes transcription of messenger RNA (mRNA) and certain small nuclear RNAs (snRNAs). |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q6T8E9 | CCNT1 | Cyclin-T1 | Bos taurus (Bovine) | SS |
| Q8HXN7 | CCNT1 | Cyclin-T1 | Pan troglodytes (Chimpanzee) | SS |
| P24863 | CCNC | Cyclin-C | Homo sapiens (Human) | PR |
| P51946 | CCNH | Cyclin-H | Homo sapiens (Human) | PR |
| O60563 | CCNT1 | Cyclin-T1 | Homo sapiens (Human) | EV |
| Q8N1B3 | CCNQ | Cyclin-Q | Homo sapiens (Human) | PR |
| Q9QWV9 | Ccnt1 | Cyclin-T1 | Mus musculus (Mouse) | SS |
| O88874 | Ccnk | Cyclin-K | Mus musculus (Mouse) | PR |
| Q2QQS5 | CYCT1-1 | Cyclin-T1-4 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z7H3 | CYCT1_2 | Cyclin-T1-2 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAC5 | CYCT1-3 | Cyclin-T1-3 | Oryza sativa subsp japonica (Rice) | PR |
| Q8GYM6 | CYCT1-4 | Cyclin-T1-4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q503D6 | ccnq | Cyclin-Q | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKENKENSSP | SVTSANLDHT | KPCWYWDKKD | LAHTPSQLEG | LDPATEARYR | REGARFIFDV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GTRLGLHYDT | LATGIIYFHR | FYMFHSFKQF | PRYVTGACCL | FLAGKVEETP | KKCKDIIKTA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RSLLNDVQFG | QFGDDPKEEV | MVLERILLQT | IKFDLQVEHP | YQFLLKYAKQ | LKGDKNKIQK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LVQMAWTFVN | DSLCTTLSLQ | WEPEIIAVAV | MYLAGRLCKF | EIQEWTSKPM | YRRWWEQFVQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DVPVDVLEDI | CHQILDLYSQ | GKQQMPHHTP | HQLQQPPSLQ | PTPQVPQVQQ | SQPSQSSEPS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| QPQQKDPQQP | AQQQQPAQQP | KKPSPQPSSP | RQVKRAVVVS | PKEENKAAEP | PPPKIPKIET |
| 370 | 380 | 390 | 400 | 410 | 420 |
| THPPLPPAHP | PPDRKPPLAA | ALGEAEPPGP | VDATDLPKVQ | IPPPAHPAPV | HQPPPLPHRP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| PPPPPSSYMT | GMSTTSSYMS | GEGYQSLQSM | MKTEGPSYGA | LPPAYGPPAH | LPYHPHVYPP |
| 490 | 500 | 510 | 520 | 530 | 540 |
| NPPPPPVPPP | PASFPPPAIP | PPTPGYPPPP | PTYNPNFPPP | PPRLPPTHAV | PPHPPPGLGL |
| 550 | 560 | 570 | |||
| PPASYPPPAV | PPGGQPPVPP | PIPPPGMPPV | GGLGRAAWMR |