O75351
Gene name |
VPS4B |
Protein name |
Vacuolar protein sorting-associated protein 4B |
Names |
Cell migration-inducing gene 1 protein, Suppressor of K(+) transport growth defect 1, Protein SKD1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9525 |
EC number |
3.6.4.6: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
Descriptions
(Annotation based on sequence homology with Q9UN37)
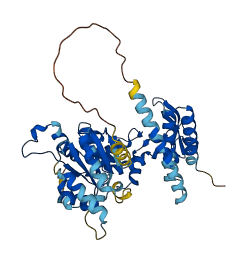
VPS4 proteins are AAA+ ATPases required to form multivesicular bodies, release viral particles, and complete cytokinesis. VPS4 proteins act by disassembling ESCRT-III heteropolymers during or after their proposed function in membrane scission. Deleting the N-terminal MIT domain and adjacent linker from VPS4A increases both basal and liposome-enhanced ATPase activity, indicating that these elements play a role in autoinhibiting VPS4A until it encounters ESCRT-III polymers. The interactions between acidic ESCRT-III residues and sequences in VPS4A, in particular in the linker connecting the MIT and AAA+ domains, are involved in regulating the intrinsic autoinhibition of the enzyme.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

220 variants for O75351
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs775341025 CA8986651 |
3 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs373658563 CA301783721 |
4 | T>I | No |
ClinGen Ensembl |
|
|
rs769719797 CA8986650 |
5 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA402642369 rs1200425372 |
6 | P>T | No |
ClinGen TOPMed |
|
|
rs539290608 CA402642355 |
7 | N>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA8986648 rs539290608 |
7 | N>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs539290608 CA402642353 |
7 | N>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA402642344 rs1369275222 |
8 | L>F | No |
ClinGen TOPMed |
|
|
CA402641644 rs1348257237 |
11 | A>V | No |
ClinGen gnomAD |
|
|
rs142770219 CA8986613 |
14 | L>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA402641595 rs1364478574 |
16 | S>G | No |
ClinGen TOPMed gnomAD |
|
|
CA301774908 rs889609154 |
17 | K>T | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 19 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1468344635 CA402641513 |
23 | K>E | No |
ClinGen gnomAD |
|
|
rs77284385 CA301774903 |
23 | K>M | No |
ClinGen Ensembl |
|
|
CA8986610 rs760728919 |
24 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA402641500 rs1204784944 |
24 | A>V | No |
ClinGen TOPMed |
|
|
CA301774881 rs1008460201 |
26 | N>D | No |
ClinGen TOPMed gnomAD |
|
|
CA8986607 rs747766401 |
28 | E>A | No |
ClinGen ExAC gnomAD |
|
|
COSM3821662 rs771753062 CA8986608 |
28 | E>K | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA8986606 rs778733915 |
29 | E>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778733915 CA402641469 |
29 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA301774875 rs112622461 |
34 | Y>H | No |
ClinGen Ensembl |
|
|
rs1284114900 CA402641420 COSM1324531 |
36 | H>R | ovary Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA402641409 rs1264129728 |
38 | V>L | No |
ClinGen gnomAD |
|
|
rs1344431459 CA402641393 |
40 | Y>C | No |
ClinGen gnomAD |
|
|
CA402641359 rs1317631687 |
45 | V>I | No |
ClinGen gnomAD |
|
|
CA402641327 rs1183371398 |
47 | Y>* | No |
ClinGen gnomAD |
|
|
CA402641313 rs1460310540 |
49 | A>V | No |
ClinGen gnomAD |
|
|
CA8986583 rs746333097 |
56 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA8986582 rs781678987 |
57 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA8986581 rs17688948 VAR_023385 |
58 | I>M | induces thermal instability [UniProt] | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1233871143 CA402641238 |
60 | A>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1315570182 CA402641226 |
62 | C>S | No |
ClinGen gnomAD |
|
|
CA402641224 rs1245148080 |
62 | C>Y | No |
ClinGen gnomAD |
|
|
CA8986577 rs572345457 |
73 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 75 | Y>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs75197917 CA301773779 |
76 | L>M | No |
ClinGen ExAC gnomAD |
|
|
rs750332149 CA8986574 |
78 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs767570340 CA8986573 |
80 | E>A | No |
ClinGen ExAC gnomAD |
|
|
CA8986570 rs763711021 |
83 | A>T | No |
ClinGen ExAC |
|
|
rs932945721 CA301773751 |
83 | A>V | No |
ClinGen TOPMed |
|
|
rs775240653 CA402641001 CA402641003 |
84 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA8986569 rs762482134 |
84 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA402641008 rs1568089202 |
84 | Q>P | No |
ClinGen Ensembl |
|
|
rs1319824000 CA402640983 |
86 | P>S | No |
ClinGen gnomAD |
|
|
CA8986565 rs532105479 |
90 | G>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA8986564 rs560222347 |
92 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs747569589 CA8986562 |
93 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs774790929 CA8986561 |
95 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA301771145 rs976152185 |
101 | D>G | No |
ClinGen TOPMed |
|
|
rs771991314 CA8986542 |
102 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA301771128 rs956770613 |
104 | G>W | No |
ClinGen TOPMed |
|
| TCGA novel | 107 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 108 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA402640413 rs1187082863 |
109 | D>G | No |
ClinGen gnomAD |
|
|
rs1263202433 CA402640416 |
109 | D>H | No |
ClinGen gnomAD |
|
|
CA402640391 rs1342965466 |
112 | E>G | No |
ClinGen TOPMed |
|
|
rs1141024 CA8986540 |
114 | K>R | No |
ClinGen ExAC gnomAD |
|
| rs750318908 | 114 | K>R | Variant assessed as Somatic; 4.719e-05 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1141024 CA402640376 |
114 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA402640361 rs1205179492 |
116 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1205274888 CA402640354 |
117 | Q>P | No |
ClinGen gnomAD |
|
|
CA402640254 rs1382565047 |
122 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
rs775333614 CA301768340 |
123 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs202058899 CA8986520 |
126 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8986521 rs768899063 |
126 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA402640221 rs1387781185 |
128 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs373315578 CA402640220 |
128 | R>P | No |
ClinGen ESP gnomAD |
|
|
COSM3422270 rs373315578 CA301768324 |
128 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP NCI-TCGA gnomAD |
|
rs1445149607 CA402640216 |
129 | P>T | No |
ClinGen gnomAD |
|
|
rs1367720947 CA402640208 |
130 | N>S | No |
ClinGen gnomAD |
|
|
CA8986519 rs780446635 |
131 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs770792561 CA8986518 |
132 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777493409 CA8986516 |
133 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA402640177 rs758341819 |
134 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA8986514 rs201158795 |
135 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs142699030 CA8986512 COSM438308 |
136 | V>I | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA gnomAD |
|
rs753435564 CA8986511 |
137 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs983145250 CA301768302 |
139 | L>H | No |
ClinGen gnomAD |
|
|
rs766028023 CA8986510 |
140 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA402640118 rs1408828233 |
144 | E>A | No |
ClinGen gnomAD |
|
|
CA402640107 rs1304398410 |
146 | L>M | No |
ClinGen gnomAD |
|
|
rs969107427 CA301768294 |
150 | V>G | No |
ClinGen Ensembl |
|
|
rs148042910 CA8986509 |
150 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA402640069 rs1367238668 |
152 | L>M | No |
ClinGen TOPMed |
|
| TCGA novel | 153 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA402640049 rs1455212232 |
155 | K>Q | No |
ClinGen gnomAD |
|
|
CA402640042 rs1302362203 |
156 | F>L | No |
ClinGen TOPMed |
|
|
CA8986505 rs775031234 |
158 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA8986504 rs768807383 |
159 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs752055600 CA8986486 |
165 | T>A | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 174 | G>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs910450361 CA301766256 |
175 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA8986479 rs771848778 |
182 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1369809114 CA402639859 |
182 | Y>H | No |
ClinGen TOPMed |
|
|
rs1319658987 CA402639839 |
185 | K>E | No |
ClinGen gnomAD |
|
|
CA402639815 rs1166762346 |
188 | A>V | No |
ClinGen TOPMed |
|
|
rs747947691 CA8986478 |
189 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA402639811 rs747947691 |
189 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM78767 rs1408021325 CA402639788 |
192 | N>K | ovary [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA8986477 rs774202246 |
193 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 194 | S>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8986476 rs768570547 |
194 | S>L | No |
ClinGen ExAC gnomAD |
|
| rs369884834 | 198 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 198 | S>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs960455655 CA301766229 |
199 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA8986475 rs772659865 |
199 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA402639690 rs755611326 |
204 | L>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755611326 CA8986473 |
204 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA402639666 rs1342434212 |
206 | S>C | No |
ClinGen TOPMed |
|
|
CA402639655 rs1462989562 |
207 | K>R | No |
ClinGen gnomAD |
|
| TCGA novel | 213 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs749187337 CA8986456 |
214 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA402639576 rs1383576196 |
214 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA402639556 rs1206175838 |
215 | L>Q | No |
ClinGen gnomAD |
|
|
CA402639544 rs1568085412 |
217 | K>M | No |
ClinGen Ensembl |
|
|
CA402639517 rs1202987143 |
221 | Q>E | No |
ClinGen gnomAD |
|
|
rs1161902773 CA402639506 |
222 | L>P | No |
ClinGen TOPMed |
|
|
rs745375968 CA8986453 |
225 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1377257364 CA402639461 |
229 | S>P | No |
ClinGen gnomAD |
|
|
CA8986452 rs780823682 |
233 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 236 | I>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8986451 rs149336699 |
240 | C>Y | No |
ClinGen ESP ExAC gnomAD |
|
|
rs746705982 CA8986450 |
241 | G>D | No |
ClinGen ExAC |
|
| TCGA novel | 246 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA402639305 rs138933226 |
251 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs138933226 CA8986445 |
251 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs372889875 CA8986444 |
252 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs753852708 COSM438306 CA8986443 |
252 | R>H | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA402639270 rs1178624815 |
256 | T>M | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 258 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA402639228 rs1244309366 |
262 | M>I | No |
ClinGen TOPMed |
|
|
rs756212918 CA8986417 |
264 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs1176173997 CA402639188 |
267 | V>I | No |
ClinGen TOPMed |
|
|
rs752422242 CA8986413 |
269 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA301765207 rs912112346 |
270 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1414070200 CA402639135 |
274 | V>A | No |
ClinGen Ensembl |
|
|
rs1599357886 CA402639131 |
275 | L>P | No |
ClinGen Ensembl |
|
|
CA8986410 rs371815146 |
280 | I>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs895709121 CA301765197 |
280 | I>V | No |
ClinGen Ensembl |
|
|
rs770516367 CA8986409 |
283 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA402639067 rs770516367 |
283 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs760270615 CA8986408 |
288 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs760759039 CA8986391 |
295 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs772832718 CA8986390 |
301 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA8986388 rs761335205 |
303 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA402638922 rs1371855334 |
303 | P>S | No |
ClinGen gnomAD |
|
|
rs768849402 CA8986386 |
304 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1189302789 CA402638910 |
305 | A>S | No |
ClinGen gnomAD |
|
|
rs1424631496 CA402638907 |
305 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA8986385 rs749668835 |
306 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA8986384 rs775900762 |
308 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1275420534 CA402638886 CA402638885 |
309 | M>I | No |
ClinGen gnomAD |
|
|
rs531693634 CA301763205 |
309 | M>T | No |
ClinGen gnomAD |
|
|
CA402638890 rs1599356735 |
309 | M>V | No |
ClinGen Ensembl |
|
|
CA402638865 rs1205129731 |
312 | L>P | No |
ClinGen gnomAD |
|
|
rs980286737 CA301763193 |
315 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
rs776365854 COSM989460 CA301763175 |
322 | T>M | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs757387117 CA8986380 |
323 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1308071182 CA402638789 |
324 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs747150958 CA8986379 |
325 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8986376 rs753679036 |
327 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA8986377 rs150331543 |
327 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs752058625 CA301763079 |
341 | I>K | No |
ClinGen gnomAD |
|
|
CA402638641 rs1234832861 |
346 | R>C | No |
ClinGen TOPMed |
|
|
rs1202244873 CA402638638 |
346 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA402638615 rs1264015297 |
350 | M>L | No |
ClinGen gnomAD |
|
|
CA8986369 rs539348167 |
353 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA402638573 rs1408619377 |
356 | V>L | No |
ClinGen gnomAD |
|
|
rs140180968 CA8986367 |
360 | T>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA402638539 rs1325512539 |
361 | H>R | No |
ClinGen gnomAD |
|
|
CA402638541 rs1185922964 |
361 | H>Y | No |
ClinGen TOPMed |
|
|
CA8986329 rs200235369 COSM1711456 |
366 | R>C | skin [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs952229759 CA301760094 |
366 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA402638489 rs1414046021 COSM564097 |
367 | G>R | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 368 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8986326 rs369063057 |
368 | P>S | No |
ClinGen ESP ExAC gnomAD |
|
|
CA402638473 rs1171095126 |
369 | S>F | No |
ClinGen gnomAD |
|
|
CA8986324 rs757925293 |
370 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs147130607 CA8986323 |
370 | R>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA8986322 rs147130607 COSM264665 |
370 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
CA402638454 rs1474232084 |
373 | P>S | No |
ClinGen gnomAD |
|
|
rs1238908521 CA402638446 |
374 | N>S | No |
ClinGen gnomAD |
|
|
CA8986320 rs754213173 |
375 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs142404608 CA8986319 |
376 | L>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs965934959 CA301760016 |
377 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs773617268 CA8986317 |
380 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs761210420 CA8986318 |
380 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs201406128 CA8986316 |
381 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs553400769 CA8986315 |
382 | T>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA301759949 rs1033915095 |
385 | S>C | No |
ClinGen TOPMed |
|
|
CA8986313 rs140107443 |
385 | S>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1447548931 CA402638379 |
386 | P>S | No |
ClinGen gnomAD |
|
|
CA402638372 rs1271988113 |
387 | G>R | No |
ClinGen TOPMed |
|
|
rs1205435046 CA402638358 |
389 | P>A | No |
ClinGen TOPMed |
|
|
rs146639296 CA8986311 |
392 | I>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA301759891 rs112699327 |
400 | P>S | No |
ClinGen Ensembl |
|
| TCGA novel | 403 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs746914721 CA8986309 |
408 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8986308 rs144476720 |
410 | S>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA301759867 rs747235022 |
411 | M>L | No |
ClinGen gnomAD |
|
|
rs1010609426 CA301759860 |
411 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
rs139544128 CA402638187 |
412 | S>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs139544128 CA8986288 |
412 | S>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs765072823 CA301757364 |
416 | R>Q | No |
ClinGen gnomAD |
|
|
CA301757359 rs1022701364 |
418 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA8986286 rs748129460 |
420 | N>K | No |
ClinGen ExAC gnomAD |
|
|
CA402638134 rs1293312191 |
421 | T>A | No |
ClinGen gnomAD |
|
| TCGA novel | 423 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8986284 rs754512421 |
425 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748915944 CA8986283 |
426 | N>D | No |
ClinGen ExAC |
|
|
CA402638102 rs1322194152 |
426 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1011747661 CA301757341 |
427 | E>K | No |
ClinGen TOPMed |
|
| TCGA novel | 429 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1207854390 CA402638020 |
437 | T>A | No |
ClinGen TOPMed |
|
|
CA402637988 rs1174587685 |
441 | G>D | No |
ClinGen gnomAD |
|
|
CA8986281 rs755866565 |
442 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA8986279 rs768043383 |
444 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8986280 rs768043383 |
444 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
No associated diseases with O75351
7 regional properties for O75351
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | AAA+ ATPase domain | 166 - 302 | IPR003593 |
| domain | ATPase, AAA-type, core | 170 - 300 | IPR003959 |
| conserved_site | ATPase, AAA-type, conserved site | 272 - 291 | IPR003960 |
| domain | MIT domain | 4 - 82 | IPR007330 |
| domain | Spastin/Vps4, C-terminal | 381 - 441 | IPR015415 |
| domain | AAA ATPase, AAA+ lid domain | 325 - 357 | IPR041569 |
| domain | Vacuolar protein sorting-associated protein 4, MIT domain | 6 - 81 | IPR045253 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.6 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
14 GO annotations of cellular component
| Name | Definition |
|---|---|
| ATPase complex | A protein complex which is capable of ATPase activity. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| Flemming body | A cell part that is the central region of the midbody characterized by a gap in alpha-tubulin staining. It is a dense structure of antiparallel microtubules from the central spindle in the middle of the intercellular bridge. |
| late endosome membrane | The lipid bilayer surrounding a late endosome. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| nuclear pore | A protein complex providing a discrete opening in the nuclear envelope of a eukaryotic cell, where the inner and outer nuclear membranes are joined. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| spindle pole | Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| identical protein binding | Binding to an identical protein or proteins. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein-containing complex binding | Binding to a macromolecular complex. |
32 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome maturation | Removal of PI3P and Atg8/LC3 after the closure of the phagophore and before the fusion with the endosome/lysosome (e.g. mammals and insects) or vacuole (yeast), and that very likely destabilizes other Atg proteins and thus enables their efficient dissociation and recycling. |
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cholesterol transport | The directed movement of cholesterol, cholest-5-en-3-beta-ol, into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| endosomal transport | The directed movement of substances mediated by an endosome, a membrane-bounded organelle that carries materials enclosed in the lumen or located in the endosomal membrane. |
| endosome to lysosome transport via multivesicular body sorting pathway | The directed movement of substances from endosomes to lysosomes by a pathway in which molecules are sorted into multivesicular bodies, which then fuse with the lysosome. |
| ESCRT III complex disassembly | The disaggregation of an ESCRT III complex into its constituent components. |
| late endosomal microautophagy | The autophagy process by which cytosolic proteins targeted for degradation are tagged with a chaperone and are directly transferred into and degraded in a late endosomal compartment. |
| late endosome to lysosome transport via multivesicular body sorting pathway | The directed movement of substances from late endosomes to lysosomes by a pathway in which molecules are sorted into multivesicular bodies, which then fuse with the lysosome. |
| macroautophagy | The major inducible pathway for the general turnover of cytoplasmic constituents in eukaryotic cells, it is also responsible for the degradation of active cytoplasmic enzymes and organelles during nutrient starvation. Macroautophagy involves the formation of double-membrane-bounded autophagosomes which enclose the cytoplasmic constituent targeted for degradation in a membrane-bounded structure. Autophagosomes then fuse with a lysosome (or vacuole) releasing single-membrane-bounded autophagic bodies that are then degraded within the lysosome (or vacuole). Some types of macroautophagy, e.g. pexophagy, mitophagy, involve selective targeting of the targets to be degraded. |
| membrane fission | A process that is carried out at the cellular level which results in the separation of a single continuous membrane into two membranes. |
| midbody abscission | The process by which the midbody, the cytoplasmic bridge that connects the two prospective daughter cells, is severed at the end of mitotic cytokinesis, resulting in two separate daughter cells. |
| mitotic metaphase plate congression | The cell cycle process in which chromosomes are aligned at the metaphase plate, a plane halfway between the poles of the mitotic spindle, during mitosis. |
| multivesicular body assembly | The aggregation, arrangement and bonding together of a set of components to form a multivesicular body, a type of late endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body sorting pathway | A vesicle-mediated transport process in which transmembrane proteins are ubiquitylated to facilitate their entry into luminal vesicles of multivesicular bodies (MVBs); upon subsequent fusion of MVBs with lysosomes or vacuoles, the cargo proteins are degraded. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of exosomal secretion | Any process that stops, prevents or reduces the frequency, rate or extent of exosomal secretion. |
| nuclear membrane reassembly | The reformation of the nuclear membranes following their breakdown in the context of a normal process. |
| nucleus organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the nucleus. |
| plasma membrane repair | The resealing of a cell plasma membrane after cellular wounding due to, for instance, mechanical stress. |
| positive regulation of centriole elongation | Any process that activates or increases the frequency, rate or extent of centriole elongation. |
| positive regulation of exosomal secretion | Any process that activates or increases the frequency, rate or extent of exosomal secretion. |
| positive regulation of G2/M transition of mitotic cell cycle | Any signalling pathway that activates or increases the activity of a cell cycle cyclin-dependent protein kinase to modulate the switch from G2 phase to M phase of the mitotic cell cycle. |
| potassium ion transport | The directed movement of potassium ions (K+) into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| protein depolymerization | The process in which protein polymers, compounds composed of a large number of component monomers, are broken down. Depolymerization occurs by the successive removal of monomers from an existing poly- or oligomeric protein. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of centrosome duplication | Any process that modulates the frequency, rate or extent of centrosome duplication. Centrosome duplication is the replication of a centrosome, a structure comprised of a pair of centrioles and peri-centriolar material from which a microtubule spindle apparatus is organized. |
| regulation of mitotic spindle assembly | Any process that modulates the frequency, rate or extent of mitotic spindle assembly. |
| response to lipid | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipid stimulus. |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | The chemical reactions and pathways resulting in the breakdown of a protein or peptide covalently tagged with ubiquitin, via the multivesicular body (MVB) sorting pathway; ubiquitin-tagged proteins are sorted into MVBs, and delivered to a lysosome/vacuole for degradation. |
| ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway | The chemical reactions and pathways resulting in the breakdown of a protein or peptide, via the multivesicular body (MVB) sorting pathway; proteins are sorted into MVBs, and delivered to a lysosome/vacuole for degradation. This process is independent of ubiquitination. |
| viral budding from plasma membrane | A viral budding that starts with formation of a membrane curvature in the host plasma membrane. |
| viral budding via host ESCRT complex | Viral budding which uses a host ESCRT protein complex, or complexes, to mediate the budding process. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P52917 | VPS4 | Vacuolar protein sorting-associated protein 4 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q0VD48 | VPS4B | Vacuolar protein sorting-associated protein 4B | Bos taurus (Bovine) | SS |
| Q9UN37 | VPS4A | Vacuolar protein sorting-associated protein 4A | Homo sapiens (Human) | EV |
| O75449 | KATNA1 | Katanin p60 ATPase-containing subunit A1 | Homo sapiens (Human) | PR |
| Q9UBP0 | SPAST | Spastin | Homo sapiens (Human) | PR |
| A6NCM1 | IQCA1L | IQ and AAA domain-containing protein 1-like | Homo sapiens (Human) | PR |
| Q86XH1 | IQCA1 | Dynein regulatory complex protein 11 | Homo sapiens (Human) | PR |
| Q6PIW4 | FIGNL1 | Fidgetin-like protein 1 | Homo sapiens (Human) | PR |
| P46467 | Vps4b | Vacuolar protein sorting-associated protein 4B | Mus musculus (Mouse) | PR |
| Q8VEJ9 | Vps4a | Vacuolar protein sorting-associated protein 4A | Mus musculus (Mouse) | PR |
| Q793F9 | Vps4a | Vacuolar protein sorting-associated protein 4A | Rattus norvegicus (Rat) | PR |
| D0FH76 | VPS4 | Vacuolar protein sorting-associated protein 4 | Bombyx mori (Silk moth) | SS |
| Q9ZNT0 | SKD1 | Protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSSTSPNLQK | AIDLASKAAQ | EDKAGNYEEA | LQLYQHAVQY | FLHVVKYEAQ | GDKAKQSIRA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KCTEYLDRAE | KLKEYLKNKE | KKAQKPVKEG | QPSPADEKGN | DSDGEGESDD | PEKKKLQNQL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QGAIVIERPN | VKWSDVAGLE | GAKEALKEAV | ILPIKFPHLF | TGKRTPWRGI | LLFGPPGTGK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SYLAKAVATE | ANNSTFFSIS | SSDLVSKWLG | ESEKLVKNLF | QLARENKPSI | IFIDEIDSLC |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GSRSENESEA | ARRIKTEFLV | QMQGVGVDND | GILVLGATNI | PWVLDSAIRR | RFEKRIYIPL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PEPHARAAMF | KLHLGTTQNS | LTEADFRELG | RKTDGYSGAD | ISIIVRDALM | QPVRKVQSAT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| HFKKVRGPSR | ADPNHLVDDL | LTPCSPGDPG | AIEMTWMDVP | GDKLLEPVVS | MSDMLRSLSN |
| 430 | 440 | ||||
| TKPTVNEHDL | LKLKKFTEDF | GQEG |