O60479
Gene name |
DLX3 |
Protein name |
Homeobox protein DLX-3 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1747 |
EC number |
|
Protein Class |
|
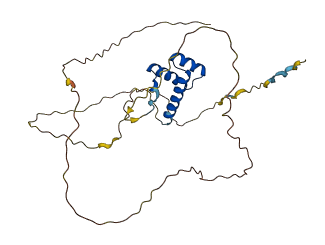
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for O60479
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4XRS | X-ray | 350 A | G/I | 131-186 | PDB |
| AF-O60479-F1 | Predicted | AlphaFoldDB |
254 variants for O60479
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000379760 RCV003165780 rs147702169 RCV000308787 CA8641120 |
28 | D>Y | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA8641106 RCV002522986 RCV000348560 rs773002632 |
47 | Q>R | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV002250427 CA8641086 rs772972332 |
88 | Y>C | Uterine leiomyoma [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
rs1598155105 RCV000984955 |
114 | E>missing | Tricho-dento-osseous syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1555617226 CA400175474 RCV000585785 |
159 | R>L | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000009641 RCV000009640 RCV001851767 rs387906406 |
188 | Y>missing | Tricho-dento-osseous syndrome Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000009639 RCV001563406 rs387906405 |
191 | G>missing | Tricho-dento-osseous syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000415464 RCV000585754 RCV001198203 rs1057518764 |
192 | E>missing | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism Amelogenesis imperfecta [ClinVar] | Yes |
ClinVar dbSNP |
|
rs146899668 CA8640961 RCV000373161 |
233 | P>L | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs142563930 RCV000308429 CA8640957 RCV001122595 |
237 | Y>C | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA8640950 RCV001128311 rs3744539 RCV002070495 |
246 | D>H | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001128310 rs1906092898 |
249 | N>T | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinVar dbSNP |
|
CA202358 rs151180891 RCV000906711 RCV000177226 RCV001128309 |
278 | G>R | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
COSM166520 rs886053135 RCV000264466 CA10640032 |
282 | N>K | Hypomaturation-hypoplastic amelogenesis imperfecta with taurodontism haematopoietic_and_lymphoid_tissue [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar TOPMed dbSNP gnomAD |
|
rs771831301 CA8641126 |
2 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1245425748 CA400179669 |
3 | G>V | No |
ClinGen gnomAD |
|
|
rs1349729555 CA400179660 |
5 | F>L | No |
ClinGen gnomAD |
|
|
rs1202513576 CA400179649 |
6 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA8641125 rs759271131 |
7 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA400179625 rs1411084054 |
7 | R>L | No |
ClinGen gnomAD |
|
|
rs1272935628 CA400179573 |
11 | S>C | No |
ClinGen TOPMed |
|
|
rs1370825778 CA400179570 |
11 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1206296718 CA400179564 |
11 | S>R | No |
ClinGen TOPMed |
|
|
CA400179568 rs1370825778 |
11 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
CA8641124 rs773905208 |
12 | I>S | No |
ClinGen ExAC gnomAD |
|
|
CA291509168 rs968374138 |
15 | D>N | No |
ClinGen gnomAD |
|
| TCGA novel | 17 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8641123 rs770570954 |
19 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8641122 rs748763551 |
19 | S>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777353435 CA8641121 |
21 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA400179382 rs1305411736 |
22 | C>R | No |
ClinGen gnomAD |
|
|
rs1021348057 CA291509151 |
23 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
CA400179334 rs866749104 |
24 | A>E | No |
ClinGen TOPMed |
|
|
CA400179341 rs1167022227 |
24 | A>S | No |
ClinGen TOPMed |
|
|
CA291509145 COSM1493932 rs866749104 |
24 | A>V | kidney [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA400179320 rs1430225377 |
25 | G>C | No |
ClinGen TOPMed |
|
|
rs1330607709 CA400179270 |
27 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA400179287 rs1233717069 |
27 | K>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 29 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs758662303 CA291509135 |
31 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs758662303 CA8641116 |
31 | T>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 33 | P>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs753928550 CA8641112 |
33 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757430047 CA8641114 |
33 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs757430047 CA8641113 |
33 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs756092425 CA8641110 |
34 | E>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400179148 rs756092425 |
34 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA400179126 rs1275496183 |
35 | S>F | No |
ClinGen TOPMed |
|
|
CA400179139 rs1215564672 |
35 | S>T | No |
ClinGen TOPMed |
|
|
CA291509052 rs1031628106 |
36 | S>P | No |
ClinGen Ensembl |
|
| TCGA novel | 36 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA400179102 rs1209951312 |
37 | V>I | No |
ClinGen TOPMed |
|
|
rs1209951312 CA400179100 |
37 | V>L | No |
ClinGen TOPMed |
|
|
rs1433855303 CA400179040 |
39 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA400178997 rs1332407095 |
41 | G>C | No |
ClinGen gnomAD |
|
|
CA400178890 rs1462760577 |
45 | A>G | No |
ClinGen TOPMed |
|
|
rs767400459 CA8641109 |
45 | A>P | No |
ClinGen ExAC gnomAD |
|
|
CA8641108 rs767400459 |
45 | A>T | Variant assessed as Somatic; 0.0001387 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1186733485 CA400178859 |
46 | P>H | No |
ClinGen TOPMed |
|
|
CA400178845 rs1168907985 |
47 | Q>* | No |
ClinGen TOPMed gnomAD |
|
|
CA400178780 rs1422722224 |
48 | H>R | No |
ClinGen TOPMed |
|
|
CA8641105 rs765952812 |
50 | Y>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs199578857 CA8641104 |
52 | S>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 53 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA291509024 rs967300082 |
54 | Q>L | No |
ClinGen TOPMed gnomAD |
|
|
CA8641102 rs769410123 |
55 | P>H | No |
ClinGen ExAC gnomAD |
|
|
rs1477172516 CA400178501 |
55 | P>T | No |
ClinGen gnomAD |
|
|
rs1194067869 CA400178436 |
56 | Y>C | No |
ClinGen gnomAD |
|
|
CA400178333 rs1464253088 |
59 | T>M | No |
ClinGen gnomAD |
|
|
CA291509014 rs368052798 |
60 | V>M | No |
ClinGen Ensembl |
|
|
CA400178273 rs1210504684 |
61 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA400178188 rs1426285240 |
63 | Y>C | No |
ClinGen gnomAD |
|
|
rs1319264267 CA400178198 |
63 | Y>H | No |
ClinGen TOPMed |
|
|
CA400178055 rs1216922938 |
67 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA8641098 rs779327348 |
68 | Q>H | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 71 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8641096 rs757474298 |
72 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA291508957 rs1048462149 |
73 | G>R | No |
ClinGen TOPMed |
|
|
CA8641095 rs749489409 |
75 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs368628839 CA8641094 |
76 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400177842 rs1319216822 |
76 | G>S | No |
ClinGen gnomAD |
|
|
rs368628839 CA291508946 |
76 | G>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1346250778 CA400177817 |
77 | T>R | No |
ClinGen gnomAD |
|
|
CA8641092 rs376068784 |
78 | G>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs767457790 CA8641091 |
79 | A>P | No |
ClinGen ExAC gnomAD |
|
|
rs1178869273 CA400177779 |
79 | A>V | No |
ClinGen gnomAD |
|
|
CA400177709 rs1448712346 |
83 | K>N | No |
ClinGen gnomAD |
|
| TCGA novel | 83 | K>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs930075852 CA291508921 |
84 | S>A | No |
ClinGen Ensembl |
|
|
CA8641090 rs754879050 |
84 | S>L | No |
ClinGen ExAC gnomAD |
|
|
rs766183369 CA8641088 |
86 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA8641087 rs761483777 |
87 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1443620648 CA400177594 COSM1384134 |
89 | G>R | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA8641085 RCV000884755 rs764946982 |
90 | A>S | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
CA8641083 rs146270612 |
93 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8641081 rs746369270 |
95 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
CA8641082 rs772519409 |
95 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs774623884 CA8641080 |
96 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs771296590 CA8641079 |
97 | A>E | No |
ClinGen ExAC gnomAD |
|
|
rs771296590 CA400177467 |
97 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1445981166 CA400177432 |
99 | R>G | No |
ClinGen gnomAD |
|
|
rs777991740 CA8641077 |
99 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777991740 CA8641078 |
99 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756272555 CA8641076 |
100 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1477606750 CA400177410 |
100 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA291508867 rs34931821 |
101 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781328319 CA400177366 |
102 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs781328319 CA8641074 |
102 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs1413043704 CA400177353 |
103 | L>P | No |
ClinGen gnomAD |
|
|
CA291508851 rs1041310386 |
104 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA400177299 rs1598155614 |
106 | Q>P | No |
ClinGen Ensembl |
|
|
rs1257609206 CA400177270 |
108 | P>A | No |
ClinGen gnomAD |
|
|
rs751496563 CA8641072 |
108 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1257609206 CA400177271 |
108 | P>T | No |
ClinGen gnomAD |
|
|
CA8641046 rs536983977 |
110 | S>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA400176201 rs1159121642 |
112 | K>E | No |
ClinGen gnomAD |
|
|
CA8641044 rs367547505 |
113 | E>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8641045 rs367547505 |
113 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs773624545 CA8641043 |
113 | E>V | No |
ClinGen ExAC gnomAD |
|
|
CA8641042 rs770122605 |
114 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA400176138 rs1470088693 |
115 | P>A | No |
ClinGen gnomAD |
|
|
rs1313269426 CA400176134 |
115 | P>L | No |
ClinGen Ensembl |
|
|
CA8641041 rs762066110 |
118 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA400176082 rs1208645623 |
118 | E>G | No |
ClinGen gnomAD |
|
|
CA291507797 rs968302616 |
118 | E>K | No |
ClinGen TOPMed |
|
|
rs200244480 CA8641040 |
120 | R>H | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs200244480 CA8641039 |
120 | R>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1242176157 CA400176067 |
120 | R>S | No |
ClinGen TOPMed |
|
|
CA8641038 rs747128275 |
121 | M>K | No |
ClinGen ExAC gnomAD |
|
|
rs1444817378 CA400176044 |
121 | M>L | No |
ClinGen TOPMed |
|
|
rs747128275 CA400176036 |
121 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs1444817378 CA400176045 |
121 | M>V | No |
ClinGen TOPMed |
|
|
CA291507770 rs1048822264 |
124 | G>A | No |
ClinGen Ensembl |
|
|
CA8641036 rs141785743 |
126 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8641037 rs141785743 |
126 | P>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs973375129 CA400175880 |
129 | V>F | No |
ClinGen TOPMed gnomAD |
|
|
rs973375129 CA291507714 |
129 | V>I | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 129 | V>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 131 | K>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA400175848 rs1386984617 |
132 | P>S | No |
ClinGen gnomAD |
|
|
CA400175822 rs1315166585 |
134 | T>K | No |
ClinGen gnomAD |
|
|
CA400175821 rs1315166585 |
134 | T>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA8641029 rs752320234 |
139 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs912828322 CA291507693 |
141 | L>V | No |
ClinGen Ensembl |
|
|
rs767002078 CA8641028 |
142 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1396074381 CA400175746 |
142 | A>V | No |
ClinGen gnomAD |
|
|
CA8641027 rs758953105 |
143 | A>G | No |
ClinGen ExAC TOPMed |
|
|
rs1188264013 CA400175736 |
143 | A>S | No |
ClinGen gnomAD |
|
|
CA8641025 rs765696358 |
145 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA8641026 rs750898874 |
145 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
rs1205897217 CA400175703 |
146 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1483058354 CA400175700 |
146 | R>H | No |
ClinGen gnomAD |
|
|
rs1205897217 CA400175707 |
146 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1254261748 CA400175648 |
149 | Q>R | No |
ClinGen gnomAD |
|
|
CA8641024 rs762264445 |
150 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs764385314 CA8641022 |
157 | P>L | No |
ClinGen ExAC |
|
|
rs1226144367 CA400175512 |
157 | P>S | No |
ClinGen gnomAD |
|
|
rs1296056573 CA400175500 |
158 | E>* | No |
ClinGen TOPMed gnomAD |
|
|
CA400175504 rs1296056573 |
158 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1314401417 CA400175432 |
162 | L>V | No |
ClinGen gnomAD |
|
|
CA400175421 rs1434784421 |
163 | A>T | No |
ClinGen gnomAD |
|
|
rs1341334320 CA400175405 |
165 | Q>H | No |
ClinGen TOPMed |
|
|
CA400175402 rs1201252049 |
166 | L>V | No |
ClinGen TOPMed |
|
|
rs774450949 CA8641017 |
168 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA400175382 rs1420553846 |
169 | T>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA400175380 rs1375088888 |
170 | Q>E | No |
ClinGen gnomAD |
|
| TCGA novel | 172 | Q>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 174 | K>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1598154436 CA400174808 |
179 | N>T | No |
ClinGen Ensembl |
|
|
rs1567913379 COSM1178609 CA400174796 |
180 | R>C | prostate [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
COSM280419 rs762788082 CA8640993 |
180 | R>H | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
| TCGA novel | 181 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1386740142 CA400174782 |
181 | R>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 184 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs769444646 CA8640991 |
186 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780848971 CA8640990 |
188 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs780848971 CA8640989 |
188 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
CA400174625 rs779537851 |
191 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8640986 rs779537851 |
191 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs145991664 CA8640987 CA400174635 |
191 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400174622 rs779537851 |
191 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA291506411 rs71379389 |
192 | E>K | No |
ClinGen Ensembl |
|
|
CA400174600 rs1457367845 |
192 | E>V | No |
ClinGen TOPMed |
|
|
rs1413495760 CA400174547 |
194 | P>L | No |
ClinGen gnomAD |
|
|
rs1198031977 CA400174508 |
198 | S>G | No |
ClinGen gnomAD |
|
|
CA8640983 rs200085639 |
200 | N>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA8640982 rs756453905 |
204 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs1225161834 CA400174301 |
205 | M>T | No |
ClinGen gnomAD |
|
|
rs1265830146 CA400174311 |
205 | M>V | No |
ClinGen gnomAD |
|
|
CA400174144 rs1425414184 |
211 | P>L | No |
ClinGen TOPMed |
|
|
rs767857178 CA8640980 |
212 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs1369437201 CA400174102 |
213 | P>L | No |
ClinGen gnomAD |
|
|
rs376543839 CA8640977 COSM980894 |
214 | A>T | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA400174006 rs773054749 CA8640975 |
216 | W>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1402690715 CA400173978 |
218 | T>P | No |
ClinGen gnomAD |
|
| TCGA novel | 219 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA291506285 rs895851859 |
220 | S>F | No |
ClinGen Ensembl |
|
|
CA291506303 rs544889964 |
220 | S>P | No |
ClinGen 1000Genomes gnomAD |
|
|
CA8640973 rs575862443 |
221 | H>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA291506281 rs575862443 |
221 | H>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs556078415 CA291506265 |
222 | S>P | No |
ClinGen 1000Genomes gnomAD |
|
|
rs768367638 CA8640971 |
223 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs542746094 CA291506263 |
223 | T>P | No |
ClinGen 1000Genomes gnomAD |
|
|
CA291506235 rs1055790164 |
224 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs746639367 CA8640970 |
224 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA400173842 rs745331311 |
225 | A>P | No |
ClinGen ExAC gnomAD |
|
|
rs745331311 CA8640967 |
225 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA8640966 rs373291908 |
228 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8640965 rs138972403 |
228 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA400173808 rs138972403 |
228 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs146899668 CA400173725 |
233 | P>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs766585893 COSM294316 CA8640959 |
234 | P>L | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA400173651 rs1184757152 |
238 | S>N | No |
ClinGen TOPMed |
|
|
rs765093320 CA8640956 |
240 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
rs761739041 CA8640955 |
241 | P>A | No |
ClinGen ExAC |
|
|
rs373417517 CA8640953 |
242 | S>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8640954 rs373417517 |
242 | S>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs760418490 CA8640952 |
244 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs3744539 CA291506085 |
246 | D>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA291506075 rs958060656 |
247 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs745373565 CA8640949 |
247 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773905882 CA8640948 |
248 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400173569 rs1296718765 |
250 | S>C | No |
ClinGen gnomAD |
|
|
rs1296718765 CA400173568 |
250 | S>F | No |
ClinGen gnomAD |
|
| TCGA novel | 251 | W>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs781719320 CA8640945 |
252 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
rs1216680558 CA400173547 |
253 | H>R | No |
ClinGen gnomAD |
|
|
rs1032099378 CA400173543 |
254 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
CA291506033 rs1032099378 COSM1750126 |
254 | A>T | Variant assessed as Somatic; 0.0 impact. urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA400173538 rs1357396789 |
255 | Q>* | No |
ClinGen gnomAD |
|
|
rs376076832 CA8640943 |
256 | N>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs376076832 CA400173529 |
256 | N>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 258 | S>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8640942 rs538517320 |
263 | Q>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA400173466 rs1410302608 |
265 | Q>* | No |
ClinGen gnomAD |
|
|
rs1412850299 CA400173455 |
266 | P>R | No |
ClinGen gnomAD |
|
|
CA400173429 rs1257535240 |
270 | A>V | No |
ClinGen gnomAD |
|
|
rs765349414 CA8640939 |
271 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs140502073 CA8640937 |
272 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8640936 rs372350880 |
273 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs775288058 CA8640934 |
274 | H>L | No |
ClinGen ExAC |
|
|
CA8640933 rs368296464 |
275 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400173387 rs151180891 |
278 | G>W | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8640931 rs770534977 |
279 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA291505907 rs895819196 |
280 | P>L | No |
ClinGen gnomAD |
|
|
CA400173376 rs1299782255 |
280 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1598154199 CA400173363 |
282 | N>S | No |
ClinGen Ensembl |
|
|
CA291505893 rs777301972 |
283 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs777301972 CA8640929 |
283 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA400173354 rs1158032945 |
284 | G>E | No |
ClinGen gnomAD |
|
|
CA400173356 rs888123487 CA291505879 |
284 | G>R | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 285 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV000350210 rs148713638 CA8640928 |
285 | A>T | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs747341304 CA8640927 |
287 | Y>C | No |
ClinGen ExAC gnomAD |
2 associated diseases with O60479
[MIM: 190320]: Trichodentoosseous syndrome (TDO)
An autosomal dominant disease characterized by curly kinky hair at birth, enamel hypoplasia, taurodontism, thickening of cortical bones and variable expression of craniofacial morphology. {ECO:0000269|PubMed:9467018}. Note=The disease is caused by variants affecting the gene represented in this entry.
[MIM: 104510]: Amelogenesis imperfecta 4 (AI4)
An autosomal dominant defect of enamel formation associated with enlarged pulp chambers. Enamel is thin, teeth are small and widely spaced. {ECO:0000269|PubMed:15666299}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal dominant disease characterized by curly kinky hair at birth, enamel hypoplasia, taurodontism, thickening of cortical bones and variable expression of craniofacial morphology. {ECO:0000269|PubMed:9467018}. Note=The disease is caused by variants affecting the gene represented in this entry.
- An autosomal dominant defect of enamel formation associated with enlarged pulp chambers. Enamel is thin, teeth are small and widely spaced. {ECO:0000269|PubMed:15666299}. Note=The disease is caused by variants affecting the gene represented in this entry.
6 regional properties for O60479
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Helix-turn-helix motif | 158 - 183 | IPR000047 |
| domain | Homeobox domain | 127 - 191 | IPR001356 |
| conserved_site | Homeobox, conserved site | 162 - 185 | IPR017970 |
| domain | Homeobox domain, metazoa | 151 - 162 | IPR020479-1 |
| domain | Homeobox domain, metazoa | 166 - 185 | IPR020479-2 |
| domain | Distal-less-like homeobox protein, N-terminal domain | 27 - 106 | IPR022135 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| blood vessel development | The process whose specific outcome is the progression of a blood vessel over time, from its formation to the mature structure. The blood vessel is the vasculature carrying blood. |
| BMP signaling pathway | The series of molecular signals initiated by the binding of a member of the BMP (bone morphogenetic protein) family to a receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| epithelial cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of an epithelial cell, any of the cells making up an epithelium. |
| hair cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a hair cell. |
| hair follicle cell proliferation | The multiplication or reproduction of hair follicle cells, resulting in the expansion of a cell population. |
| hair follicle morphogenesis | The process in which the anatomical structures of the hair follicle are generated and organized. |
| odontoblast differentiation | The process in which a relatively unspecialized cell of neural crest origin acquires the specialized features of an odontoblast, a cell on the outer surface of the dental pulp whose biological function is the creation of dentin. |
| odontogenesis of dentin-containing tooth | The process whose specific outcome is the progression of a dentin-containing tooth over time, from its formation to the mature structure. A dentin-containing tooth is a hard, bony organ borne on the jaw or other bone of a vertebrate, and is composed mainly of dentin, a dense calcified substance, covered by a layer of enamel. |
| placenta development | The process whose specific outcome is the progression of the placenta over time, from its formation to the mature structure. The placenta is an organ of metabolic interchange between fetus and mother, partly of embryonic origin and partly of maternal origin. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P50577 | DLX5 | Homeobox protein DLX-5 | Gallus gallus (Chicken) | PR |
| Q3C1V8 | BSX | Brain-specific homeobox protein homolog | Homo sapiens (Human) | PR |
| P56179 | DLX6 | Homeobox protein DLX-6 | Homo sapiens (Human) | PR |
| P70397 | Dlx6 | Homeobox protein DLX-6 | Mus musculus (Mouse) | PR |
| P70396 | Dlx5 | Homeobox protein DLX-5 | Mus musculus (Mouse) | PR |
| Q64205 | Dlx3 | Homeobox protein DLX-3 | Mus musculus (Mouse) | PR |
| P50575 | Dlx5 | Homeobox protein DLX-5 | Rattus norvegicus (Rat) | PR |
| Q9FN29 | ATHB-52 | Homeobox-leucine zipper protein ATHB-52 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q98875 | dlx1a | Homeobox protein Dlx1a | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q98878 | dlx4b | Homeobox protein Dlx4b | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| P50574 | dlx2a | Homeobox protein Dlx2a | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q01702 | dlx3b | Homeobox protein Dlx3b | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSGSFDRKLS | SILTDISSSL | SCHAGSKDSP | TLPESSVTDL | GYYSAPQHDY | YSGQPYGQTV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NPYTYHHQFN | LNGLAGTGAY | SPKSEYTYGA | SYRQYGAYRE | QPLPAQDPVS | VKEEPEAEVR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MVNGKPKKVR | KPRTIYSSYQ | LAALQRRFQK | AQYLALPERA | ELAAQLGLTQ | TQVKIWFQNR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RSKFKKLYKN | GEVPLEHSPN | NSDSMACNSP | PSPALWDTSS | HSTPAPARSQ | LPPPLPYSAS |
| 250 | 260 | 270 | 280 | ||
| PSYLDDPTNS | WYHAQNLSGP | HLQQQPPQPA | TLHHASPGPP | PNPGAVY |