O54785
Gene name |
Limk2 |
Protein name |
LIM domain kinase 2 |
Names |
LIMK-2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:16886 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
468-511 (Activation loop from InterPro)
Target domain |
331-608 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
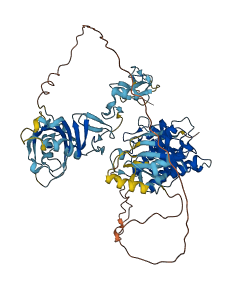
Autoinhibited structure

Activated structure

2 structures for O54785
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2YUB | NMR | - | A | 142-246 | PDB |
| AF-O54785-F1 | Predicted | AlphaFoldDB |
26 variants for O54785
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389077422 | 10 | W>C | No | EVA | |
| rs3389114028 | 14 | G>S | No | EVA | |
| rs3401797249 | 15 | C>S | No | EVA | |
| rs3389125517 | 28 | T>I | No | EVA | |
| rs3389136789 | 39 | R>Q | No | EVA | |
| rs3389132026 | 107 | D>E | No | EVA | |
| rs3389137919 | 114 | V>M | No | EVA | |
| rs3389125453 | 123 | K>T | No | EVA | |
| rs3389125497 | 212 | V>I | No | EVA | |
| rs3401940699 | 214 | T>P | No | EVA | |
| rs3389136820 | 246 | Q>H | No | EVA | |
| rs26884113 | 248 | R>Q | No | EVA | |
| rs3389149660 | 257 | M>K | No | EVA | |
| rs3389103013 | 259 | S>N | No | EVA | |
| rs26884114 | 263 | T>M | No | EVA | |
| rs3389147291 | 284 | L>Q | No | EVA | |
| rs3389111235 | 313 | E>D | No | EVA | |
| rs3389142992 | 324 | Q>H | No | EVA | |
| rs3389139183 | 338 | G>R | No | EVA | |
| rs26884119 | 368 | E>D | No | EVA | |
| rs3401890305 | 419 | S>T | No | EVA | |
| rs3389102957 | 500 | R>C | No | EVA | |
| rs3389143020 | 557 | G>V | No | EVA | |
| rs3389140286 | 598 | E>K | No | EVA | |
| rs217822317 | 621 | D>E | No | EVA | |
| rs3389111287 | 627 | S>G | No | EVA |
No associated diseases with O54785
6 regional properties for O54785
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 331 - 608 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 333 - 599 | IPR001245 |
| domain | PDZ domain | 152 - 239 | IPR001478 |
| domain | Zinc finger, LIM-type | 10 - 70 | IPR001781-1 |
| domain | Zinc finger, LIM-type | 71 - 131 | IPR001781-2 |
| binding_site | Protein kinase, ATP binding site | 337 - 360 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cis-Golgi network | The network of interconnected tubular and cisternal structures located at the convex side of the Golgi apparatus, which abuts the endoplasmic reticulum. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| mitotic spindle | A spindle that forms as part of mitosis. Mitotic and meiotic spindles contain distinctive complements of proteins associated with microtubules. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| astral microtubule organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of astral microtubules, any of the spindle microtubules that radiate in all directions from the spindle poles. |
| cornea development in camera-type eye | The progression of the cornea over time, from its formation to the mature structure. The cornea is the transparent structure that covers the anterior of the eye. |
| establishment of vesicle localization | The directed movement of a vesicle to a specific location. |
| head development | The biological process whose specific outcome is the progression of a head from an initial condition to its mature state. The head is the anterior-most division of the body. |
| negative regulation of cilium assembly | Any process that stops, prevents or reduces the frequency, rate or extent of cilium assembly. |
| positive regulation of protein localization to nucleus | Any process that activates or increases the frequency, rate or extent of protein localization to nucleus. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
27 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9Y616 | IRAK3 | Interleukin-1 receptor-associated kinase 3 | Homo sapiens (Human) | PR |
| P53667 | LIMK1 | LIM domain kinase 1 | Homo sapiens (Human) | PR |
| Q96S53 | TESK2 | Dual specificity testis-specific protein kinase 2 | Homo sapiens (Human) | PR |
| P53671 | LIMK2 | LIM domain kinase 2 | Homo sapiens (Human) | PR |
| Q8K4B2 | Irak3 | Interleukin-1 receptor-associated kinase 3 | Mus musculus (Mouse) | PR |
| Q8CFA1 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Mus musculus (Mouse) | PR |
| Q8VCT9 | Tesk2 | Dual specificity testis-specific protein kinase 2 | Mus musculus (Mouse) | PR |
| Q4QQS0 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Rattus norvegicus (Rat) | PR |
| Q924U5 | Tesk2 | Dual specificity testis-specific protein kinase 2 | Rattus norvegicus (Rat) | PR |
| Q9ASQ5 | CRCK3 | Calmodulin-binding receptor-like cytoplasmic kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STF0 | LECRKS3 | Receptor like protein kinase S.3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCZ4 | FER | Receptor-like protein kinase FERONIA | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LYN6 | At3g56050 | Probable inactive receptor-like protein kinase At3g56050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94C25 | At5g20050 | Probable receptor-like protein kinase At5g20050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLW0 | At5g24010 | Probable receptor-like protein kinase At5g24010 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LTC0 | PBL19 | Probable serine/threonine-protein kinase PBL19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FIL7 | CRCK1 | Calmodulin-binding receptor-like cytoplasmic kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAALAGDEAW | RCRGCGTYVP | LSQRLYRTAN | EAWHGSCFRC | SECQESLTNW | YYEKDGKLYC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| HKDYWAKFGE | FCHGCSLLMT | GPAMVAGEFK | YHPECFACMS | CKVIIEDGDA | YALVQHATLY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CGKCHNEVVL | APMFERLSTE | SVQDQLPYSV | TLISMPATTE | CRRGFSVTVE | SASSNYATTV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QVKEVNRMHI | SPNNRNAIHP | GDRILEINGT | PVRTLRVEEV | EDAIKQTSQT | LQLLIEHDPV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PQRLDQLRLD | ARLPPHMQST | GHTLMLSTLD | TKENQEGTLR | RRSLRRSNSI | SKSPGPSSPK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EPLLLSRDIS | RSESLRCSSS | YSQQIFRPCD | LIHGEVLGKG | FFGQAIKVTH | KATGKVMVMK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ELIRCDEETQ | KTFLTEVKVM | RSLDHPNVLK | FIGVLYKDKK | LNLLTEYIEG | GTLKDFLRSV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DPFPWQQKVR | FAKGISSGMA | YLHSMCIIHR | DLNSHNCLIK | LDKTVVVADF | GLSRLIVEER |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KRPPVEKATT | KKRTLRKSDR | KKRYTVVGNP | YWMAPEMLNG | KSYDETVDVF | SFGIVLCEII |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GQVYADPDCL | PRTLDFGLNV | KLFWEKFVPT | DCPPAFFPLA | AICCKLEPES | RPAFSKLEDS |
| 610 | 620 | 630 | |||
| FEALSLFLGE | LAIPLPAELE | DLDHTVSMEY | GLTRDSPP |