O43896
Gene name |
KIF1C (KIAA0706) |
Protein name |
Kinesin-like protein KIF1C |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:10749 |
EC number |
|
Protein Class |
|
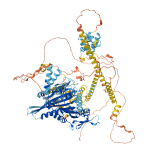
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-436 (Motor domain and neck coil) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for O43896
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2G1L | X-ray | 260 A | A | 498-599 | PDB |
| AF-O43896-F1 | Predicted | AlphaFoldDB |
1208 variants for O43896
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000684908 rs747424547 |
5 | S>L | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000645365 rs756071061 CA8318423 |
13 | R>W | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
ClinGen ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV000645367 rs1368090910 CA397342753 |
39 | N>S | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
RCV001772075 RCV003166229 RCV000803511 rs202033753 |
42 | Q>H | Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV002540253 rs771071450 RCV001768695 RCV003120666 |
51 | T>I | Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
RCV000799553 rs201085674 |
75 | R>W | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001965030 rs572156268 |
104 | S>F | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes dbSNP |
|
rs757847238 RCV000821734 |
118 | I>L | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs745851014 RCV001056498 |
129 | R>C | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001848513 rs773746672 RCV002034764 |
138 | L>V | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001391533 rs2143312682 |
149 | I>F | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs1291871672 RCV001934859 |
155 | R>G | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV003258975 RCV000799939 RCV002267025 rs199962814 |
167 | R>Q | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. Inborn genetic diseases [ClinVar, NCI-TCGA] | Yes |
ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
CA8318586 RCV000540072 rs185479618 |
167 | R>W | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV000087324 COSM1521700 COSM6147257 VAR_070937 rs587777198 RCV003421989 CA150725 |
169 | R>W | lung Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. SPAX2 [Cosmic, ClinVar, NCI-TCGA, UniProt] | Yes |
ClinGen cosmic curated NCI-TCGA Cosmic ClinVar UniProt NCI-TCGA dbSNP gnomAD |
|
RCV001093146 rs772475828 RCV001219292 |
176 | P>L | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001953284 rs1974623326 RCV003303470 |
183 | K>T | Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar TOPMed dbSNP |
|
rs752855724 RCV001330255 RCV002546376 |
190 | A>T | Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000822526 rs754938138 |
193 | A>T | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
rs2143313439 RCV001918167 |
201 | K>missing | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA16620467 RCV000485547 rs1064796693 RCV000985162 |
216 | R>C | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000687046 RCV003258918 RCV001252794 rs141189136 |
228 | R>C | Spastic ataxia 2 Microcephaly Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001004868 rs1361059674 |
233 | L>F | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV001330256 rs201655443 RCV002546377 |
234 | T>M | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. Inborn genetic diseases [ClinVar, NCI-TCGA] | Yes |
ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
rs2143317762 RCV001389057 |
243 | K>missing | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs139697370 RCV000871383 |
244 | I>M | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs2143317920 RCV001647140 |
256 | D>missing | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001864750 RCV002545883 rs150280414 |
256 | D>N | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. Inborn genetic diseases [ClinVar, NCI-TCGA] | Yes |
ClinVar 1000Genomes ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
CA8318771 RCV000488299 RCV001085446 RCV001848856 rs146872023 |
289 | Q>P | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000871153 RCV002539099 rs145650252 RCV001446098 RCV001847085 |
295 | S>L | Hereditary spastic paraplegia Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000824133 rs760721111 |
300 | Y>F | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV002538750 rs187228739 RCV001757834 |
349 | R>C | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1597846084 RCV001003621 RCV000850202 |
351 | N>missing | Intellectual disability Cerebellar ataxia [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000497507 CA8318865 RCV000516075 rs142056835 RCV001079974 RCV000487792 |
371 | A>T | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs367997542 RCV000811319 |
372 | R>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001848958 RCV002292564 CA8318875 rs79290524 RCV000553957 |
379 | A>G | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs755935386 RCV001848493 RCV002543406 RCV002543405 |
400 | A>T | Hereditary spastic paraplegia Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001295488 rs1259655283 |
421 | G>R | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV001973595 rs2143331530 |
426 | S>L | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001848494 RCV002034763 rs139663513 |
431 | T>M | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs144939400 RCV001231494 |
434 | Q>H | Spastic ataxia 2 [ClinVar] | Yes |
1000Genomes ESP ExAC TOPMed gnomAD ClinVar dbSNP |
|
RCV001452858 rs148619121 RCV002549567 RCV001847121 |
437 | P>L | Hereditary spastic paraplegia Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000699867 rs1567722733 |
441 | M>I | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001848914 RCV000521926 RCV001321550 CA8318947 rs142046798 |
449 | K>R | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002553098 rs374100485 RCV001759752 RCV001043582 |
451 | I>M | Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001235396 rs759516774 RCV000996458 |
463 | R>C | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1013580197 RCV001948387 |
540 | R>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV001979815 rs764242002 |
547 | R>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1485258329 RCV001848497 |
554 | G>R | Hereditary spastic paraplegia [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV001871113 rs1371284245 |
598 | N>K | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
rs760659714 RCV002545272 RCV001848498 |
615 | P>L | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1283601775 RCV000814248 |
616 | G>E | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV001057549 rs1975068657 |
618 | P>S | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs566736417 RCV001172109 RCV001209516 |
669 | R>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA250364 RCV001847848 RCV000191099 rs148934699 RCV000415815 |
700 | P>L | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs138935423 RCV000558940 RCV001081975 CA8319218 RCV001848789 |
702 | T>I | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs147429300 RCV002545273 RCV001848501 RCV002545274 |
719 | R>C | Hereditary spastic paraplegia Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. Inborn genetic diseases [ClinVar, NCI-TCGA] | Yes |
ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
rs587777197 RCV000087323 CA150723 |
731 | R>* | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs376385209 RCV000821598 |
739 | P>L | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001848502 rs376385209 |
739 | P>R | Hereditary spastic paraplegia [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs1453593139 RCV001941167 |
744 | M>T | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV001318630 rs767797244 |
765 | R>C | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000516023 RCV001085955 rs118037269 RCV000645370 CA8319262 |
767 | G>R | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs552440254 RCV001915186 |
768 | R>Q | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV001212450 rs377528420 |
789 | P>L | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001342537 rs764241869 |
790 | D>N | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1393424251 RCV001238953 |
793 | G>R | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
rs182610831 RCV000807662 |
810 | E>A | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001998573 rs776041349 |
815 | A>T | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs533724781 RCV002204116 |
824 | G>R | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV000492986 RCV001267982 CA645369639 rs1131690773 |
828 | A>missing | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar dbSNP |
|
RCV000645366 CA287186200 rs539082919 |
829 | E>K | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes TOPMed dbSNP gnomAD |
|
rs780729030 RCV000701825 |
841 | T>M | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs767390082 RCV001848503 |
855 | D>E | Hereditary spastic paraplegia [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001323445 rs372705324 |
856 | R>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000498097 RCV002524080 RCV001848862 CA8319324 rs143671350 RCV000820352 |
860 | A>T | Hereditary spastic paraplegia Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA8319328 RCV003409670 RCV000515870 rs146628704 RCV000809178 |
864 | R>H | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs2143389526 RCV002016143 |
866 | L>H | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs2143392214 RCV001848504 |
878 | H>R | Hereditary spastic paraplegia [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001339057 rs1297313354 |
884 | E>K | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV001998608 rs768593463 RCV003355724 RCV002267136 |
892 | P>L | Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs561679213 RCV001848505 |
892 | P>S | Hereditary spastic paraplegia [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV000692445 rs751072487 |
901 | E>K | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000535949 rs1395728872 CA397317106 |
904 | P>R | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
RCV000645368 rs758990694 CA8319368 |
907 | R>C | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA8319370 rs78970955 RCV001848769 RCV000548436 RCV000419762 |
909 | P>Q | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
COSM3680509 rs202232792 RCV000191100 RCV000859538 CA250366 RCV000516061 |
912 | R>W | Hereditary spastic paraplegia Spastic ataxia 2 large_intestine [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002048023 rs2143392621 COSM3519481 |
914 | P>S | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
NCI-TCGA Cosmic ClinVar Ensembl dbSNP |
|
RCV000996459 RCV003160133 rs200436795 |
917 | P>T | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1555571945 CA658658530 RCV000526698 |
917 | P>T | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001314928 rs200925951 |
926 | R>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001338550 rs774901685 |
940 | R>H | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
COSM4067838 RCV001848507 rs757104315 |
960 | G>R | Hereditary spastic paraplegia Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
ExAC TOPMed gnomAD NCI-TCGA Cosmic ClinVar NCI-TCGA dbSNP |
|
COSM271765 rs768222656 RCV001767719 RCV002539135 |
963 | R>C | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. large_intestine [ClinVar, NCI-TCGA, Cosmic] | Yes |
NCI-TCGA Cosmic cosmic curated ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
rs550136320 CA8319435 RCV001350123 RCV001848915 RCV000523662 |
974 | D>N | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001848508 rs2143393636 |
983 | S>R | Hereditary spastic paraplegia [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs750130519 RCV001848509 |
984 | N>K | Hereditary spastic paraplegia [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000700580 rs372031774 |
984 | N>T | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ESP TOPMed dbSNP gnomAD |
|
rs1975146105 RCV001303010 |
987 | H>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs115332491 RCV001848961 RCV000541436 CA8319446 RCV001644654 |
996 | S>N | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002541251 RCV001786869 rs774701979 |
997 | G>R | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV003298127 rs267604960 RCV000797956 |
1000 | P>L | Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000808959 COSM1384502 rs143987985 |
1002 | P>L | Spastic ataxia 2 large_intestine [ClinVar, Cosmic] | Yes |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000876192 rs185185243 RCV001847103 |
1017 | A>T | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001371602 rs2143394246 |
1017 | A>V | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001763530 rs2143394321 RCV003163854 |
1020 | P>A | Inborn genetic diseases [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
CA8319471 RCV000551616 rs369620316 |
1021 | P>L | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs372792586 RCV001057325 |
1024 | R>* | Variant assessed as Somatic; HIGH impact. Spastic ataxia 2 [NCI-TCGA, ClinVar] | Yes |
ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV000525348 RCV001090632 rs141225452 RCV001848791 CA8319474 |
1024 | R>Q | Hereditary spastic paraplegia Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs752457302 RCV002568969 RCV001547661 |
1027 | H>Q | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV002544910 RCV001849055 RCV000691418 rs62072492 RCV000996460 |
1030 | R>C | Hereditary spastic paraplegia Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs779584975 RCV000804412 |
1033 | S>F | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs201887996 RCV002066442 |
1041 | R>Q | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV003166690 RCV001300848 RCV001847234 rs200388087 |
1041 | R>W | Hereditary spastic paraplegia Spastic ataxia 2 Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001952849 rs1199087791 |
1043 | A>T | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. [ClinVar, NCI-TCGA] | Yes |
ClinVar NCI-TCGA dbSNP gnomAD |
|
rs373541138 COSM981381 RCV000810755 RCV002538076 |
1043 | A>V | Spastic ataxia 2 Variant assessed as Somatic; MODERATE impact. endometrium Inborn genetic diseases [ClinVar, NCI-TCGA, Cosmic] | Yes |
NCI-TCGA Cosmic cosmic curated ClinVar 1000Genomes ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV001301486 rs1240123302 |
1047 | Q>R | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP |
|
rs199966544 RCV002000921 |
1073 | P>S | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs1975155981 RCV001334577 |
1079 | P>H | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001895778 rs753993722 |
1080 | P>missing | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000953949 RCV000502000 rs201800868 CA8319527 |
1091 | R>H | Spastic ataxia 2 [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001370717 rs1269241991 |
1093 | A>T | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV001242669 rs1975158981 |
1098 | E>K | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001049069 rs375445356 |
1101 | A>missing | Spastic ataxia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001268867 rs1468786988 |
1 | M>I | No |
ClinVar dbSNP |
|
| rs1974545420 | 2 | A>S | No | TOPMed | |
| rs776032010 | 3 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs776032010 | 3 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs776931555 | 4 | A>G | No |
ExAC gnomAD |
|
| rs769053728 | 4 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs769053728 | 4 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs773227449 | 6 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs773227449 | 6 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs74766284 | 7 | K>E | No | Ensembl | |
| rs1261572906 | 8 | V>A | No |
TOPMed gnomAD |
|
| rs1204479297 | 12 | V>I | No | gnomAD | |
| rs1436391935 | 13 | R>Q | No |
TOPMed gnomAD |
|
| rs776354474 | 17 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs776354474 | 17 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs199632760 | 18 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs756916879 | 18 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs199632760 | 18 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1470002802 | 19 | E>K | No | gnomAD | |
| rs1974548430 | 20 | T>N | No | Ensembl | |
| rs201845629 | 21 | S>G | No |
1000Genomes ExAC gnomAD |
|
| rs1392900029 | 21 | S>R | No |
TOPMed gnomAD |
|
| rs745553914 | 21 | S>T | No |
ExAC gnomAD |
|
| rs200913968 | 22 | Q>* | No |
TOPMed gnomAD |
|
| rs200913968 | 22 | Q>E | No |
TOPMed gnomAD |
|
| rs1404468705 | 23 | D>N | No |
TOPMed gnomAD |
|
| rs757956577 | 23 | D>V | No |
ExAC gnomAD |
|
| rs1224387824 | 25 | K>R | No |
TOPMed gnomAD |
|
| rs1250920860 | 26 | C>R | No | gnomAD | |
| rs761316876 | 27 | V>M | No |
TOPMed gnomAD |
|
| rs1974549703 | 28 | V>G | No | Ensembl | |
| rs1337912749 | 28 | V>I | No |
TOPMed gnomAD |
|
| rs1974549874 | 29 | S>R | No | Ensembl | |
| TCGA novel | 30 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1308685223 | 33 | N>T | No | gnomAD | |
| rs1974550375 | 35 | T>N | No | Ensembl | |
| rs2143306396 | 36 | S>C | No | Ensembl | |
| rs1974550575 | 36 | S>P | No | TOPMed | |
| rs1974568326 | 37 | I>N | No | TOPMed | |
| rs751047597 | 37 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1974568402 | 38 | I>F | No | Ensembl | |
| rs377337240 | 38 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed |
| rs867838488 | 40 | P>L | No | Ensembl | |
| rs905447373 | 44 | K>E | No |
TOPMed gnomAD |
|
| rs1288378400 | 45 | D>N | No |
TOPMed gnomAD |
|
| rs1347751013 | 46 | A>V | No | gnomAD | |
| rs1321412412 | 47 | P>L | No |
TOPMed gnomAD |
|
| rs748695798 | 47 | P>S | No |
ExAC gnomAD |
|
| rs748695798 | 47 | P>T | No |
ExAC gnomAD |
|
| rs778176215 | 49 | S>N | No |
ExAC TOPMed gnomAD |
|
|
rs1430555089 COSM981338 |
49 | S>R | endometrium [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs575703589 | 51 | T>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs771071450 | 51 | T>N | No |
ExAC gnomAD |
|
| rs1258435489 | 54 | Y>C | No | gnomAD | |
| rs1974570020 | 54 | Y>H | No |
TOPMed gnomAD |
|
| rs1974570301 | 57 | W>C | No | Ensembl | |
| COSM1384495 | 58 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776684520 | 59 | H>Q | No |
ExAC gnomAD |
|
| rs555256113 | 61 | S>L | No |
1000Genomes ExAC gnomAD |
|
| rs1355100100 | 61 | S>P | No | TOPMed | |
| rs762688180 | 62 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs773096943 | 62 | T>S | No | ExAC | |
| rs773854559 | 63 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs759293239 | 64 | D>E | No |
ExAC gnomAD |
|
| rs887921341 | 65 | P>L | No | TOPMed | |
| rs997594436 | 65 | P>S | No |
TOPMed gnomAD |
|
| rs766946486 | 68 | A>V | No |
ExAC gnomAD |
|
|
COSM417451 rs1186425348 |
70 | Q>* | Variant assessed as Somatic; HIGH impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1376160688 | 73 | V>L | No | gnomAD | |
| rs1376160688 | 73 | V>M | No | gnomAD | |
| rs1282096656 | 74 | Y>S | No | gnomAD | |
| rs201085674 | 75 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1224990876 | 75 | R>Q | No | gnomAD | |
| rs1974584015 | 77 | I>V | No | gnomAD | |
| rs1306748912 | 78 | G>R | No | gnomAD | |
| rs1017623562 | 79 | E>K | No | TOPMed | |
| rs747410405 | 80 | E>D | No |
ExAC gnomAD |
|
| COSM981339 | 83 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1974584605 | 84 | H>Y | No | Ensembl | |
| rs779390411 | 85 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs746070005 | 88 | G>D | No |
ExAC gnomAD |
|
| rs1006736815 | 89 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs758655441 | 90 | N>D | No |
ExAC gnomAD |
|
| rs535190367 | 90 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 90 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747110877 | 91 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs747110877 | 91 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1168760173 | 92 | C>Y | No | gnomAD | |
| rs769814577 | 93 | I>M | No |
ExAC gnomAD |
|
| rs138283031 | 95 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1356574441 | 95 | A>V | No | gnomAD | |
| rs749298736 | 98 | Q>H | No |
ExAC gnomAD |
|
| rs368782086 | 99 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1275146785 | 100 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1974586539 | 101 | A>D | No | Ensembl | |
| rs1370798795 | 101 | A>T | No | gnomAD | |
|
rs1974586605 RCV001093145 |
102 | G>A | No |
ClinVar Ensembl dbSNP |
|
| rs572156268 | 104 | S>Y | No | 1000Genomes | |
| rs767179480 | 106 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1342553983 | 106 | T>P | No | gnomAD | |
| rs1269729856 | 108 | M>V | No | Ensembl | |
| rs1292427447 | 109 | G>R | No |
TOPMed gnomAD |
|
| rs774939814 | 110 | R>* | No |
ExAC gnomAD |
|
| rs1489891485 | 110 | R>Q | No | gnomAD | |
| TCGA novel | 111 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760142761 | 112 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs763639870 | 113 | P>L | No |
ExAC gnomAD |
|
| rs1383923359 | 119 | V>A | No |
TOPMed gnomAD |
|
| rs372339577 | 119 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs372339577 | 119 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1426072494 | 120 | P>S | No | gnomAD | |
| rs1350098672 | 122 | L>F | No | gnomAD | |
| rs1974607924 | 123 | C>* | No | TOPMed | |
| rs369631181 | 123 | C>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs757101369 | 125 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs778878542 | 126 | L>F | No |
ExAC gnomAD |
|
| rs974955088 | 127 | F>L | No | TOPMed | |
| rs1974608417 | 127 | F>L | No | TOPMed | |
| rs150602558 | 129 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs745851014 | 129 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs775959015 | 130 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs775959015 | 130 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs775959015 | 130 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs761420923 | 136 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs769231942 | 137 | Q>P | No |
ExAC gnomAD |
|
| rs578027417 | 139 | S>C | No |
1000Genomes TOPMed gnomAD |
|
|
rs1974609642 COSM336078 |
140 | Y>H | lung [Cosmic] | No |
cosmic curated Ensembl |
| rs1444228500 | 142 | V>A | No | gnomAD | |
| COSM4930378 | 142 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3819984 | 143 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1974619632 | 144 | V>L | No | Ensembl | |
| rs1055603292 | 145 | S>R | No |
TOPMed gnomAD |
|
| rs200106622 | 147 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1268773364 | 147 | M>V | No |
TOPMed gnomAD |
|
| rs748568140 | 148 | E>* | No | Ensembl | |
| rs756493686 | 150 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1371154380 | 150 | Y>N | No | gnomAD | |
| rs756493686 | 150 | Y>S | No |
ExAC TOPMed gnomAD |
|
| rs1240181000 | 151 | C>R | No | gnomAD | |
| rs528902778 | 153 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM981344 rs528902778 |
153 | R>Q | endometrium [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs764164742 | 153 | R>W | No |
ExAC TOPMed gnomAD |
|
|
rs1291871672 COSM1318474 |
155 | R>* | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1392886784 | 155 | R>Q | No |
TOPMed gnomAD |
|
| rs1363516013 | 158 | L>W | No | gnomAD | |
| rs1299067327 | 161 | K>E | No | gnomAD | |
| rs1359365653 | 161 | K>M | No | gnomAD | |
| COSM3712384 | 161 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV000523836 rs747834094 CA8318584 |
163 | R>Q | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
COSM3519464 rs780907252 |
163 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA |
| rs1974621753 | 164 | G>C | No |
TOPMed gnomAD |
|
| rs755727352 | 164 | G>V | No |
ExAC gnomAD |
|
| COSM3795815 | 165 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1259197761 | 168 | V>L | No | gnomAD | |
| rs1188956517 | 169 | R>Q | No | gnomAD | |
| TCGA novel | 170 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1162917513 | 170 | E>K | No |
TOPMed gnomAD |
|
| COSM981347 | 172 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1974622599 | 173 | I>V | No | Ensembl | |
| rs772475828 | 176 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs746419418 | 176 | P>S | No |
ExAC gnomAD |
|
| rs372500741 | 178 | V>L | No |
TOPMed gnomAD |
|
| rs372500741 | 178 | V>M | No |
TOPMed gnomAD |
|
| rs764242879 | 179 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1382440375 | 180 | D>N | No | gnomAD | |
| rs1974623326 | 183 | K>R | No | TOPMed | |
| rs776654657 | 186 | V>A | No |
ExAC gnomAD |
|
| rs777596594 | 188 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
|
COSM4067813 rs1204108834 |
189 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs1204108834 | 189 | Y>S | No | TOPMed | |
| rs752855724 | 190 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1330790473 | 193 | A>V | No |
TOPMed gnomAD |
|
| rs1974624406 | 196 | M>I | No | TOPMed | |
| rs767276837 | 196 | M>R | No |
ExAC gnomAD |
|
| rs767276837 | 196 | M>T | No |
ExAC gnomAD |
|
| rs558474743 | 196 | M>V | No | gnomAD | |
| rs748880540 | 197 | D>N | No | TOPMed | |
| TCGA novel | 199 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1183578623 | 200 | N>H | No | gnomAD | |
| rs1256771819 | 200 | N>S | No | gnomAD | |
| rs755749267 | 203 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1974624895 | 203 | R>W | No | TOPMed | |
| rs1974628193 | 206 | A>G | No | Ensembl | |
| rs781604565 | 207 | A>V | No |
ExAC gnomAD |
|
| rs938481858 | 210 | M>L | No |
TOPMed gnomAD |
|
| rs938481858 | 210 | M>V | No |
TOPMed gnomAD |
|
| rs748307324 | 211 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1974628533 | 212 | E>* | No | TOPMed | |
| rs770019961 | 212 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs770019961 | 212 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1226149924 | 214 | S>T | No | gnomAD | |
| rs199548525 | 215 | S>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs915801478 | 216 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs760757050 | 220 | V>I | No |
ExAC gnomAD |
|
| rs1974629429 | 222 | T>I | No | gnomAD | |
| rs1597842237 | 222 | T>P | No | Ensembl | |
| rs1974629484 | 223 | I>V | No | Ensembl | |
| rs776580434 | 224 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1974629852 | 227 | Q>H | No | Ensembl | |
| rs764827083 | 227 | Q>R | No |
ExAC gnomAD |
|
| rs142513387 | 228 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| COSM4067814 | 229 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs370106692 | 229 | C>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs370106692 | 229 | C>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1301993103 | 229 | C>Y | No | gnomAD | |
| rs755485274 | 230 | H>R | No |
ExAC gnomAD |
|
| rs1597842271 | 230 | H>Y | No |
TOPMed gnomAD |
|
| rs1293728035 | 231 | D>E | No | gnomAD | |
| rs1394933073 | 231 | D>H | No |
TOPMed gnomAD |
|
| rs1974630903 | 235 | G>A | No | TOPMed | |
| rs1974630903 | 235 | G>E | No | TOPMed | |
| rs201464566 | 237 | D>G | No | 1000Genomes | |
|
COSM3370808 rs777832656 |
238 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs756742766 | 238 | S>T | No | Ensembl | |
| rs2143317729 | 241 | V>I | No | 1000Genomes | |
| rs1974657114 | 242 | S>G | No | gnomAD | |
| rs917561706 | 243 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs750671441 | 243 | K>T | No |
ExAC gnomAD |
|
| rs1370070467 | 245 | S>G | No |
TOPMed gnomAD |
|
| rs1974657646 | 250 | A>G | No | Ensembl | |
| rs1408072376 | 252 | S>N | No |
TOPMed gnomAD |
|
| rs780225611 | 254 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs751686969 | 255 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs749235969 | 256 | D>E | No |
ExAC gnomAD |
|
|
rs903875496 COSM706867 |
257 | S>F | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs770752941 | 258 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs1974658621 | 258 | S>L | No |
TOPMed gnomAD |
|
| rs1974658971 | 259 | G>A | No | Ensembl | |
| rs1974658852 | 259 | G>R | No | TOPMed | |
| rs1974659029 | 260 | A>S | No | Ensembl | |
| rs778681371 | 261 | R>G | No |
ExAC gnomAD |
|
| rs745461601 | 261 | R>P | No |
ExAC gnomAD |
|
| rs745461601 | 261 | R>Q | No |
ExAC gnomAD |
|
| rs778681371 | 261 | R>W | No |
ExAC gnomAD |
|
| COSM436922 | 262 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1303501499 | 262 | G>V | No |
TOPMed gnomAD |
|
| rs755067749 | 263 | M>I | No | Ensembl | |
|
RCV002211382 rs557126775 |
264 | R>C | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs774939825 | 264 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs774939825 | 264 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs774939825 | 264 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1974659626 | 266 | K>Q | No |
TOPMed gnomAD |
|
| rs908801559 | 269 | A>T | No | TOPMed | |
| rs776012115 | 269 | A>V | No |
ExAC gnomAD |
|
| rs1437960066 | 270 | N>S | No |
TOPMed gnomAD |
|
| rs1296881328 | 271 | I>M | No |
TOPMed gnomAD |
|
| rs1974664265 | 271 | I>T | No | Ensembl | |
| rs1974664200 | 271 | I>V | No | Ensembl | |
| COSM3519467 | 272 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs939497458 | 272 | N>S | No |
TOPMed gnomAD |
|
| rs1974664611 | 276 | T>I | No | Ensembl | |
| rs1356614549 | 277 | T>A | No |
TOPMed gnomAD |
|
| rs770447158 | 277 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1974665003 | 278 | L>P | No | gnomAD | |
| rs1974665119 | 280 | K>R | No | Ensembl | |
| TCGA novel | 281 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1185527775 | 283 | S>L | No | gnomAD | |
| rs368151227 | 284 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1457719530 | 284 | A>V | No | gnomAD | |
| rs1205063803 | 286 | A>T | No |
TOPMed gnomAD |
|
| rs1974665992 | 287 | D>E | No | Ensembl | |
| rs1974665884 | 287 | D>N | No | Ensembl | |
| rs1974665946 | 287 | D>V | No | Ensembl | |
| rs1974667785 | 290 | S>P | No | Ensembl | |
| rs771325468 | 293 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs774589252 | 293 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs775821474 | 299 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs2143319943 | 299 | P>L | No | 1000Genomes | |
| COSM4901539 | 299 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760721111 | 300 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1974668508 | 302 | D>N | No | TOPMed | |
| rs1974668646 | 304 | V>L | No | TOPMed | |
| rs2143320136 | 313 | L>F | No | Ensembl | |
|
COSM161974 rs758685066 |
314 | G>A | breast [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs758685066 | 314 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs2143323281 | 315 | G>E | No | Ensembl | |
| rs376963307 | 316 | N>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs376963307 | 316 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs138321138 | 318 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs138321138 | 318 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs776654539 | 318 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs138321138 | 318 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs76817927 | 319 | T>P | No | Ensembl | |
| rs748146702 | 321 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1391848908 | 322 | I>T | No | gnomAD | |
| rs769968364 | 323 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1974682451 | 325 | L>M | No | gnomAD | |
| rs1324096379 | 329 | D>V | No | TOPMed | |
| rs1597845594 | 330 | I>S | No | Ensembl | |
| rs1974682710 | 330 | I>V | No | gnomAD | |
| rs767357962 | 331 | N>S | No |
1000Genomes ExAC gnomAD |
|
| rs1567721495 | 336 | L>R | No | Ensembl | |
| rs373741991 | 336 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs756706873 | 337 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs756706873 | 337 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1597845632 | 337 | S>R | No | Ensembl | |
| rs1332090479 | 340 | R>S | No | gnomAD | |
| rs1974689636 | 341 | Y>C | No | TOPMed | |
| rs1472804058 | 342 | A>P | No | gnomAD | |
| rs1467789736 | 343 | D>N | No |
TOPMed gnomAD |
|
| rs370727070 | 344 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs370727070 | 344 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM706864 rs539953623 |
344 | R>H | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs755325968 | 345 | T>A | No |
ExAC gnomAD |
|
| rs781606691 | 346 | K>R | No |
ExAC gnomAD |
|
| rs1974690294 | 347 | Q>* | No | Ensembl | |
| rs965335718 | 349 | R>H | No |
TOPMed gnomAD |
|
| rs187228739 | 349 | R>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1974690692 RCV001268296 |
350 | C>missing | No |
ClinVar dbSNP |
|
| rs148307470 | 350 | C>Y | No |
ESP TOPMed gnomAD |
|
| rs777671002 | 351 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs1210999379 | 351 | N>S | No | gnomAD | |
| rs749352816 | 353 | I>L | No |
ExAC gnomAD |
|
| rs749352816 | 353 | I>V | No |
ExAC gnomAD |
|
| rs771180950 | 354 | I>T | No |
ExAC gnomAD |
|
| rs1183475749 | 354 | I>V | No |
TOPMed gnomAD |
|
| rs747008418 | 356 | E>G | No |
ExAC gnomAD |
|
| rs1396825705 | 357 | D>H | No | gnomAD | |
| rs1435186505 | 358 | P>R | No | gnomAD | |
| rs1974691442 | 358 | P>S | No | Ensembl | |
| rs2143324699 | 359 | N>S | No | Ensembl | |
| rs921300827 | 360 | A>D | No |
TOPMed gnomAD |
|
| rs768497069 | 360 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs761322983 | 361 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs761322983 | 361 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs776499585 | 361 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs142056835 | 371 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs201287076 RCV000513474 CA8318866 |
372 | R>W | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1450624792 | 374 | R>Q | No |
TOPMed gnomAD |
|
| rs764691825 | 374 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs150594020 | 375 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs753131759 | 376 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs778030228 | 378 | M>I | No |
ExAC TOPMed gnomAD |
|
| COSM3819987 | 381 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs867611760 | 383 | S>P | No | Ensembl | |
| rs1974693477 | 384 | A>S | No |
TOPMed gnomAD |
|
| rs368324220 | 384 | A>V | No | Ensembl | |
| rs2143325089 | 385 | S>F | No | Ensembl | |
| rs1045112397 | 388 | E>A | No |
TOPMed gnomAD |
|
| rs1045112397 | 388 | E>G | No |
TOPMed gnomAD |
|
| rs367987211 | 389 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1974747638 | 392 | T>A | No | TOPMed | |
|
rs775831145 COSM3787374 |
392 | T>M | pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs781167716 | 393 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs755935386 | 400 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs777437767 | 401 | L>V | No |
ExAC gnomAD |
|
| rs748884069 | 403 | A>S | No |
ExAC gnomAD |
|
| rs748884069 | 403 | A>T | No |
ExAC gnomAD |
|
| rs371688297 | 403 | A>V | No |
ESP TOPMed gnomAD |
|
| rs770596601 | 404 | V>M | No |
ExAC gnomAD |
|
| rs773814610 | 406 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs773814610 | 406 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs1194761969 | 407 | P>S | No |
TOPMed gnomAD |
|
| rs2143331334 | 408 | P>R | No | Ensembl | |
| rs1240822680 | 411 | V>I | No | TOPMed | |
| rs1304629667 | 412 | S>A | No |
TOPMed gnomAD |
|
| rs1287537384 | 412 | S>L | No |
TOPMed gnomAD |
|
| rs1974748952 | 413 | P>L | No |
TOPMed gnomAD |
|
| rs1974748897 | 413 | P>S | No | Ensembl | |
| rs745355114 | 414 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs558102986 | 416 | P>L | No |
1000Genomes ExAC gnomAD |
|
| rs1347324446 | 417 | T>A | No | gnomAD | |
| rs1377147448 | 417 | T>N | No | TOPMed | |
| TCGA novel | 418 | T>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776056933 | 418 | T>I | No |
ExAC gnomAD |
|
| rs1974749459 | 419 | H>N | No | TOPMed | |
| rs2033017181 | 420 | N>Y | No | TOPMed | |
| rs761088146 | 422 | E>K | No |
ExAC gnomAD |
|
| rs571542781 | 423 | L>P | No |
1000Genomes ExAC gnomAD |
|
| rs1034923851 | 423 | L>V | No |
TOPMed gnomAD |
|
| rs1466254443 | 424 | E>D | No | Ensembl | |
| rs1183237957 | 424 | E>K | No |
TOPMed gnomAD |
|
| rs1183237957 | 424 | E>Q | No |
TOPMed gnomAD |
|
| rs990768881 | 425 | P>L | No |
TOPMed gnomAD |
|
| rs1307896810 | 425 | P>S | No |
TOPMed gnomAD |
|
|
TCGA novel rs1974750180 |
427 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1175011174 | 427 | F>Y | No | gnomAD | |
| rs750724414 | 428 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs750724414 | 428 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs765525997 | 428 | S>P | No |
ExAC TOPMed |
|
| rs905730505 | 429 | P>H | No |
TOPMed gnomAD |
|
| rs905730505 | 429 | P>R | No |
TOPMed gnomAD |
|
| rs752616609 | 429 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs777498924 | 430 | N>S | No |
ExAC gnomAD |
|
| rs777498924 | 430 | N>T | No |
ExAC gnomAD |
|
| rs2143331623 | 433 | S>P | No | Ensembl | |
| rs1282621759 | 433 | S>Y | No | gnomAD | |
| rs747499692 | 438 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1178143401 | 439 | E>K | No | gnomAD | |
| rs1974751372 | 439 | E>V | No | Ensembl | |
| rs371522240 | 440 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs777318222 | 441 | M>R | No |
ExAC gnomAD |
|
|
rs777318222 COSM4067831 |
441 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC gnomAD |
| rs1974751495 | 441 | M>V | No | Ensembl | |
| rs1395201005 | 445 | Q>* | No | gnomAD | |
| rs1395201005 | 445 | Q>E | No | gnomAD | |
| rs1448349099 | 445 | Q>P | No | gnomAD | |
| rs1974757510 | 448 | E>K | No | TOPMed | |
| rs1597849827 | 449 | K>E | No | Ensembl | |
| rs1287212818 | 450 | I>F | No | gnomAD | |
| rs746470176 | 451 | I>V | No |
ExAC gnomAD |
|
| COSM706862 | 452 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1292901745 | 452 | A>T | No | gnomAD | |
| TCGA novel | 452 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748733484 | 453 | E>Q | No |
ExAC gnomAD |
|
| rs770218338 | 454 | L>M | No |
ExAC gnomAD |
|
| rs377105023 | 454 | L>P | No |
ESP ExAC gnomAD |
|
|
rs1974758424 COSM1384497 |
456 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs771093765 | 457 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1186849916 | 458 | W>* | No | gnomAD | |
| rs774678551 | 458 | W>R | No |
ExAC gnomAD |
|
| rs1974758703 | 460 | E>K | No | gnomAD | |
| rs942506591 | 461 | K>R | No | Ensembl | |
| rs1974758845 | 462 | L>P | No |
TOPMed gnomAD |
|
| rs767647581 | 463 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1406913083 | 467 | A>S | No | gnomAD | |
| rs1406913083 | 467 | A>T | No | gnomAD | |
| rs1974759135 | 467 | A>V | No | TOPMed | |
| rs1435658114 | 477 | A>T | No |
TOPMed gnomAD |
|
| rs1974762742 | 478 | E>* | No |
TOPMed gnomAD |
|
| rs1974762795 | 479 | M>I | No | TOPMed | |
| COSM1303051 | 480 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1597850194 | 481 | V>G | No | Ensembl | |
| rs146881155 | 483 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 483 | V>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374701430 | 484 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs374701430 | 484 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs571606260 | 484 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs200862583 | 485 | E>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1173171673 | 486 | D>G | No | gnomAD | |
| rs1173171673 | 486 | D>V | No | gnomAD | |
| rs1396041841 | 488 | G>A | No | gnomAD | |
| rs1396041841 | 488 | G>E | No | gnomAD | |
| rs1597850230 | 489 | T>S | No | Ensembl | |
| rs772309791 | 490 | V>M | No |
ExAC gnomAD |
|
| rs1440871070 | 491 | G>S | No | gnomAD | |
| rs368199251 | 492 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1234402013 | 493 | F>L | No | gnomAD | |
| rs1484270853 | 496 | K>R | No |
TOPMed gnomAD |
|
| rs1974764016 | 497 | K>M | No | TOPMed | |
| rs1974912700 | 498 | T>S | No | TOPMed | |
|
rs1974912754 TCGA novel |
499 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs763492867 | 500 | H>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 500 | H>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763492867 | 500 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1382210749 | 501 | L>P | No | gnomAD | |
| rs1974912956 | 501 | L>V | No | TOPMed | |
| rs1383423752 | 502 | V>L | No | gnomAD | |
| rs140692517 | 508 | P>H | No |
ESP TOPMed gnomAD |
|
| rs140692517 | 508 | P>L | No |
ESP TOPMed gnomAD |
|
| rs761261999 | 508 | P>S | No |
ExAC gnomAD |
|
| rs535557199 | 509 | L>P | No |
1000Genomes ExAC gnomAD |
|
| rs1175360051 | 510 | M>I | No |
TOPMed gnomAD |
|
| rs1340812836 | 511 | S>T | No | gnomAD | |
| rs1974913538 | 513 | C>Y | No | Ensembl | |
|
COSM3742321 rs750941844 |
515 | L>F | liver [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs758695684 | 520 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs758695684 | 520 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1265302907 | 521 | G>D | No | TOPMed | |
| rs1206981630 | 522 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1974914280 | 523 | T>N | No | Ensembl | |
| rs1597855807 | 523 | T>P | No | Ensembl | |
| rs747822842 | 526 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs769279948 | 527 | Q>R | No |
ExAC gnomAD |
|
| rs771984501 | 530 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs771984501 | 530 | M>R | No |
ExAC TOPMed gnomAD |
|
| rs771984501 | 530 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs900236276 | 530 | M>V | No |
TOPMed gnomAD |
|
| rs1974936394 | 532 | I>M | No |
TOPMed gnomAD |
|
| rs1049225304 | 533 | K>M | No |
TOPMed gnomAD |
|
| rs1049225304 | 533 | K>R | No |
TOPMed gnomAD |
|
| rs1049225304 | 533 | K>T | No |
TOPMed gnomAD |
|
| rs766830103 | 535 | T>I | No |
ExAC gnomAD |
|
|
RCV001760811 rs960973563 |
536 | G>A | No |
ClinVar dbSNP gnomAD |
|
| rs960973563 | 536 | G>E | No | gnomAD | |
| rs759883791 | 536 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1974936822 | 537 | Q>H | No | TOPMed | |
| rs775303966 | 538 | F>L | No | gnomAD | |
| rs752955920 | 540 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1374652943 | 541 | E>D | No | gnomAD | |
| rs1465937783 | 542 | Q>* | No | gnomAD | |
| TCGA novel | 543 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1442946036 | 544 | C>F | No | gnomAD | |
| rs764242002 | 547 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs146618482 | 547 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1381930434 | 549 | I>T | No | gnomAD | |
| rs2143358695 | 549 | I>V | No | Ensembl | |
| rs1320196641 | 550 | P>R | No |
TOPMed gnomAD |
|
| rs753932855 | 550 | P>T | No |
ExAC gnomAD |
|
| TCGA novel | 551 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1219641121 | 551 | Q>H | No | gnomAD | |
| rs757438747 | 552 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs746926292 | 553 | D>H | No |
ExAC gnomAD |
|
| rs754799167 | 553 | D>V | No |
ExAC gnomAD |
|
| rs1374089924 | 557 | V>L | No |
TOPMed gnomAD |
|
| rs1975054803 | 559 | T>S | No | TOPMed | |
| rs1373271554 | 562 | P>L | No | gnomAD | |
|
RCV001767246 rs1285088031 |
562 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinVar NCI-TCGA TOPMed dbSNP gnomAD |
| rs777770224 | 563 | C>Y | No | Ensembl | |
| rs150226133 | 565 | G>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs199563353 | 565 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1975055352 | 568 | T>A | No | gnomAD | |
| rs1975055352 | 568 | T>S | No | gnomAD | |
| TCGA novel | 569 | Y>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1210596198 | 570 | V>A | No | gnomAD | |
| rs1443717935 | 570 | V>M | No | gnomAD | |
| rs1232044119 | 571 | N>Y | No | gnomAD | |
| rs370888080 | 576 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs770419005 | 577 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1423334662 | 578 | P>L | No |
TOPMed gnomAD |
|
| rs1423334662 | 578 | P>Q | No |
TOPMed gnomAD |
|
| rs1258390004 | 578 | P>S | No | gnomAD | |
| rs1258390004 | 578 | P>T | No | gnomAD | |
| rs557132499 | 579 | L>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs557132499 | 579 | L>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1597861664 | 580 | V>G | No | Ensembl | |
| rs1975056596 | 584 | G>R | No | TOPMed | |
| rs1204560902 | 586 | R>S | No |
TOPMed gnomAD |
|
| COSM4917718 | 591 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs745324466 | 594 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1384498 rs771628064 |
596 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs780359535 | 596 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1241749143 | 597 | F>L | No | gnomAD | |
| rs1476940480 | 600 | P>L | No |
TOPMed gnomAD |
|
| rs1975066273 | 603 | A>G | No | TOPMed | |
| rs1975066199 | 603 | A>T | No | TOPMed | |
| rs1161974448 | 607 | R>Q | No | gnomAD | |
| rs762096055 | 607 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1405460222 | 609 | R>G | No | gnomAD | |
| rs773503454 | 609 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs773503454 | 609 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs762882623 | 611 | V>G | No |
ExAC gnomAD |
|
| rs566159956 | 612 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1975067542 | 612 | P>S | No | gnomAD | |
| COSM3193307 | 613 | P>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1597862088 | 613 | P>R | No | Ensembl | |
| rs944512777 | 614 | P>L | No |
TOPMed gnomAD |
|
| rs1356876702 | 614 | P>T | No | gnomAD | |
| rs1195837794 | 615 | P>T | No |
TOPMed gnomAD |
|
|
COSM179043 rs1487454027 |
617 | P>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1487454027 | 617 | P>Q | No |
TOPMed gnomAD |
|
| rs753593337 | 618 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1567727743 | 622 | V>G | No | Ensembl | |
|
COSM473041 rs2143380778 |
623 | D>N | kidney Variant assessed as Somatic; MODERATE impact. large_intestine [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| TCGA novel | 624 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM981363 | 628 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs374353519 | 629 | K>R | No |
ESP ExAC gnomAD |
|
| rs2143380819 | 630 | E>G | No | Ensembl | |
| rs149354055 | 631 | L>M | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1975069539 COSM3519479 |
633 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs943680163 | 635 | Q>E | No |
TOPMed gnomAD |
|
| rs1975069700 | 636 | G>D | No |
TOPMed gnomAD |
|
| rs1304665405 | 638 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs755583443 | 641 | L>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 641 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748412191 | 645 | K>N | No |
ExAC gnomAD |
|
| rs1394085981 | 646 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1975074136 | 647 | L>P | No | Ensembl | |
| rs766079602 | 649 | D>E | No |
ExAC gnomAD |
|
| rs1332170518 | 649 | D>G | No | gnomAD | |
| rs751270593 | 651 | E>A | No |
ExAC gnomAD |
|
| rs751270593 | 651 | E>G | No |
ExAC gnomAD |
|
| rs1385381479 | 651 | E>K | No | gnomAD | |
| rs1331843012 | 652 | N>D | No | gnomAD | |
| rs932121492 | 654 | Y>H | No | Ensembl | |
| rs1320673365 | 655 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs753129039 | 655 | R>W | No |
ExAC TOPMed gnomAD |
|
|
COSM1610483 rs1199796606 |
656 | K>E | liver [Cosmic] | No |
cosmic curated gnomAD |
| rs1264412952 | 657 | E>D | No | gnomAD | |
| rs756691035 | 659 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs199538708 | 660 | E>K | No |
ExAC gnomAD |
|
| rs943732493 | 661 | A>V | No | TOPMed | |
| rs866317638 | 662 | D>H | No |
TOPMed gnomAD |
|
| rs866317638 | 662 | D>N | No |
TOPMed gnomAD |
|
| rs866317638 | 662 | D>Y | No |
TOPMed gnomAD |
|
| rs60007010 | 663 | L>F | No | Ensembl | |
| rs754186279 | 664 | L>R | No |
ExAC TOPMed gnomAD |
|
|
RCV001814415 rs2143382149 |
666 | E>* | No |
ClinVar Ensembl dbSNP |
|
| rs1975075520 | 666 | E>D | No | gnomAD | |
| rs1466117948 | 666 | E>G | No | gnomAD | |
| rs1466117948 | 666 | E>V | No | gnomAD | |
| rs1347985608 | 668 | Q>H | No | gnomAD | |
| rs1567727901 | 668 | Q>R | No | Ensembl | |
| rs1456471745 | 669 | R>* | No |
TOPMed gnomAD |
|
| rs1237589868 | 671 | Y>C | No | gnomAD | |
| rs1352308943 | 671 | Y>H | No | gnomAD | |
| rs1290636190 | 672 | A>V | No |
TOPMed gnomAD |
|
| rs757618663 | 674 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs757618663 | 674 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs1399120250 | 676 | S>N | No |
TOPMed gnomAD |
|
| rs1381514125 | 677 | G>A | No | TOPMed | |
| rs1381514125 | 677 | G>E | No | TOPMed | |
| rs1235493761 | 677 | G>R | No |
TOPMed gnomAD |
|
| rs1235493761 | 677 | G>W | No |
TOPMed gnomAD |
|
| rs551884040 | 678 | D>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs551884040 | 678 | D>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs551884040 | 678 | D>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs768630780 | 683 | R>C | No |
ExAC TOPMed gnomAD |
|
|
COSM1679413 rs1002320182 |
683 | R>H | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs777697918 | 684 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs777697918 | 684 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs777697918 | 684 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs976760309 | 684 | S>Y | No |
TOPMed gnomAD |
|
| rs1401462041 | 685 | C>S | No | gnomAD | |
| rs1333966276 | 687 | E>D | No |
TOPMed gnomAD |
|
| rs1163238446 | 687 | E>G | No |
TOPMed gnomAD |
|
| rs1975103366 | 688 | S>G | No | gnomAD | |
| rs1392339428 | 689 | W>* | No |
TOPMed gnomAD |
|
|
COSM3378252 rs749304445 |
689 | W>R | upper_aerodigestive_tract pancreas [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs2143386971 | 692 | I>M | No | Ensembl | |
| rs1168790116 | 692 | I>T | No |
TOPMed gnomAD |
|
| rs532735917 | 693 | S>F | No | Ensembl | |
| rs770835360 | 694 | S>F | No |
ExAC gnomAD |
|
| rs1426525617 | 695 | L>F | No | TOPMed | |
| rs759229352 | 696 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs756143565 | 696 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs771713573 | 698 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs771713573 | 698 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs775022402 | 698 | Q>R | No |
ExAC gnomAD |
|
| rs1975104234 | 701 | P>H | No | Ensembl | |
| rs142944883 | 703 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs142944883 | 703 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs758785693 | 704 | V>I | No |
ExAC gnomAD |
|
| rs946593293 | 707 | I>T | No |
TOPMed gnomAD |
|
| rs1975104828 | 707 | I>V | No | TOPMed | |
| rs766563607 | 710 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs904984161 | 710 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs766563607 | 710 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1975105524 | 713 | L>P | No |
TOPMed gnomAD |
|
| rs1975105753 | 717 | G>S | No | Ensembl | |
| rs1416519198 | 718 | K>N | No | gnomAD | |
| rs749212413 | 718 | K>R | No | ExAC | |
| rs147429300 | 719 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs779022340 | 719 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1975106276 | 720 | R>G | No | Ensembl | |
| rs745607170 | 721 | A>T | No |
ExAC gnomAD |
|
| rs771770679 | 721 | A>V | No |
ExAC gnomAD |
|
| rs1435397417 | 722 | P>T | No | gnomAD | |
| rs375660983 | 723 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs139701770 | 723 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1257145521 | 724 | R>G | No |
TOPMed gnomAD |
|
| rs1316030701 | 724 | R>S | No | gnomAD | |
| rs1479170495 | 724 | R>T | No | Ensembl | |
| rs1257145521 | 724 | R>W | No |
TOPMed gnomAD |
|
| rs993547611 | 725 | V>F | No |
TOPMed gnomAD |
|
| rs993547611 | 725 | V>I | No |
TOPMed gnomAD |
|
| rs761344460 | 726 | Y>C | No |
ExAC gnomAD |
|
| rs1200427899 | 726 | Y>H | No | gnomAD | |
| rs1198164297 | 727 | Q>P | No | gnomAD | |
| rs369357478 | 728 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs763310157 | 731 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs779215813 | 732 | R>C | No | gnomAD | |
| rs766718357 | 732 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1397753368 | 734 | L>Q | No | gnomAD | |
| rs1404259115 | 736 | G>C | No |
TOPMed gnomAD |
|
| rs1975107934 | 737 | K>N | No | Ensembl | |
| rs371774970 | 737 | K>R | No |
ESP ExAC gnomAD |
|
| rs755203770 | 738 | D>E | No |
ExAC gnomAD |
|
| rs1405143174 | 739 | P>S | No | gnomAD | |
| rs752781012 | 740 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs752781012 | 740 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs756282892 | 740 | R>H | No |
ExAC gnomAD |
|
|
RCV001268849 rs1975108349 |
741 | W>* | No |
ClinVar Ensembl dbSNP |
|
| rs745870744 | 742 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs758203086 | 743 | T>A | No |
ExAC gnomAD |
|
|
TCGA novel rs1975108575 |
744 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1975108662 | 745 | A>S | No | TOPMed | |
| rs746643030 | 745 | A>V | No |
ExAC gnomAD |
|
| rs1975108850 | 747 | L>P | No |
TOPMed gnomAD |
|
| rs1975108953 | 750 | Q>R | No | TOPMed | |
| rs1374164959 | 751 | A>T | No | gnomAD | |
| rs1472733846 | 751 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs371464539 | 754 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1975109202 | 754 | E>Q | No | TOPMed | |
| rs763427425 | 758 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs763427425 | 758 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1318684253 | 760 | A>V | No |
TOPMed gnomAD |
|
| rs573302051 | 761 | L>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs573302051 | 761 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1597864117 | 763 | D>A | No | Ensembl | |
| rs1246720031 | 764 | F>L | No |
TOPMed gnomAD |
|
| rs1361676959 | 764 | F>V | No | gnomAD | |
| rs767797244 | 765 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1487403605 | 765 | R>H | No |
TOPMed gnomAD |
|
| rs1597864152 | 767 | G>E | No | TOPMed | |
| rs575805660 | 768 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1369760038 | 770 | E>K | No | gnomAD | |
| rs780072767 | 772 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs369289976 | 773 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs376569688 | 773 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs780757171 | 774 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs117621046 | 776 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA TOPMed gnomAD |
| rs1453502889 | 777 | L>F | No | gnomAD | |
| rs1453502889 | 777 | L>I | No | gnomAD | |
| COSM4834642 | 778 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs988526971 | 779 | M>I | No | TOPMed | |
| rs1975111346 | 779 | M>T | No | Ensembl | |
| rs199782010 | 780 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs749770926 | 780 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs199782010 | 780 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1344742800 | 781 | E>D | No | gnomAD | |
| rs1199019425 | 783 | C>Y | No | gnomAD | |
| rs771298564 | 784 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs771298564 | 784 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM981375 rs774940396 |
784 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2087644436 | 785 | T>I | No |
TOPMed gnomAD |
|
| rs1441229659 | 786 | Y>C | No |
TOPMed gnomAD |
|
| rs772567048 | 787 | G>A | No |
ExAC gnomAD |
|
| rs772567048 | 787 | G>D | No |
ExAC gnomAD |
|
| rs1182080806 | 787 | G>S | No | gnomAD | |
| rs980675901 | 789 | P>S | No | TOPMed | |
| TCGA novel | 790 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs540945645 | 791 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2143388362 | 792 | P>A | No | Ensembl | |
| rs1248555798 | 792 | P>R | No |
TOPMed gnomAD |
|
| TCGA novel | 792 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2143388406 | 793 | G>A | No | Ensembl | |
| rs186917988 | 794 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1975112790 | 794 | D>V | No | TOPMed | |
| rs190022215 | 795 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs190022215 | 795 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs551749680 | 797 | R>K | No |
1000Genomes gnomAD |
|
| rs1324951439 | 798 | A>T | No | gnomAD | |
| rs1255684775 | 799 | V>L | No | gnomAD | |
| rs1195813496 | 801 | R>G | No |
TOPMed gnomAD |
|
| rs746521760 | 801 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs746521760 | 801 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1195813496 | 801 | R>W | No |
TOPMed gnomAD |
|
| rs1975113669 | 803 | V>I | No | TOPMed | |
| rs777508180 | 805 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1200786787 | 805 | D>N | No | gnomAD | |
| TCGA novel | 806 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1477487835 | 807 | V>I | No |
TOPMed gnomAD |
|
| rs1477487835 | 807 | V>L | No |
TOPMed gnomAD |
|
| rs201219607 | 809 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201219607 | 809 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1298767713 | 810 | E>D | No | gnomAD | |
| rs182610831 | 810 | E>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs779219604 | 810 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1390325462 | 811 | E>K | No |
TOPMed gnomAD |
|
| rs1385288886 | 812 | G>E | No |
TOPMed gnomAD |
|
| rs1975114855 | 812 | G>R | No | TOPMed | |
| rs1975115166 | 813 | G>D | No | Ensembl | |
| rs1415223968 | 813 | G>S | No |
TOPMed gnomAD |
|
| rs2143388712 | 814 | G>R | No | Ensembl | |
| TCGA novel | 816 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747413228 | 816 | G>S | No |
ExAC gnomAD |
|
| rs956991329 | 817 | S>C | No | TOPMed | |
| rs1975115825 | 817 | S>I | No | Ensembl | |
| rs1309728888 | 817 | S>R | No | gnomAD | |
| rs568266722 | 818 | G>A | No |
1000Genomes ExAC gnomAD |
|
| rs1597864446 | 821 | S>R | No | Ensembl | |
| rs1264319328 | 821 | S>T | No | gnomAD | |
| rs1487328639 | 822 | E>K | No | gnomAD | |
| rs1269418400 | 823 | E>K | No | gnomAD | |
| rs547704394 | 825 | A>T | No |
1000Genomes TOPMed gnomAD |
|
| rs1425712894 | 826 | R>* | No |
TOPMed gnomAD |
|
| rs1425712894 | 826 | R>G | No |
TOPMed gnomAD |
|
| rs761733935 | 826 | R>Q | No |
ExAC gnomAD |
|
| COSM1384500 | 827 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 828 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1367955935 | 828 | A>V | No |
TOPMed gnomAD |
|
| rs558996508 | 829 | E>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1597864516 | 830 | V>G | No | Ensembl | |
| rs767267650 | 831 | E>D | No |
ExAC gnomAD |
|
| rs1364751336 | 833 | L>F | No |
TOPMed gnomAD |
|
| rs776267022 | 834 | R>Q | No |
TOPMed gnomAD |
|
|
COSM5055603 rs575669774 |
834 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA gnomAD |
| rs763591946 | 835 | A>V | No |
ExAC gnomAD |
|
| rs756714924 | 836 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs778537068 | 837 | I>V | No |
ExAC gnomAD |
|
| rs139084469 | 838 | D>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs139084469 | 838 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs139084469 | 838 | D>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1181207786 | 839 | K>R | No |
TOPMed gnomAD |
|
| rs776716271 | 842 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs769148587 | 842 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1975119152 | 843 | I>F | No | Ensembl | |
| rs1173153721 | 845 | Q>K | No |
TOPMed gnomAD |
|
| rs1415492201 | 845 | Q>R | No | gnomAD | |
| rs769739540 | 846 | E>D | No |
ExAC gnomAD |
|
| rs1314947427 | 847 | V>A | No | gnomAD | |
| rs1314947427 | 847 | V>G | No | gnomAD | |
| rs1975119473 | 847 | V>M | No | Ensembl | |
| rs773380705 | 848 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1302779533 | 850 | Q>K | No | gnomAD | |
| rs1454134901 | 852 | S>N | No | gnomAD | |
| rs1315085304 | 853 | S>N | No | gnomAD | |
| rs1338957579 | 853 | S>R | No |
TOPMed gnomAD |
|
| rs1237475147 | 855 | D>Y | No | gnomAD | |
| rs372705324 | 856 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs554739220 | 856 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1254204034 | 857 | E>K | No | gnomAD | |
| rs1567728964 | 860 | A>G | No | Ensembl | |
| rs143671350 | 860 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1567728964 | 860 | A>V | No | Ensembl | |
| rs1597864678 | 861 | L>P | No | Ensembl | |
|
RCV001757959 rs749907095 |
862 | R>Q | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
COSM5852981 rs764833076 |
862 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1238693741 | 863 | D>E | No | gnomAD | |
|
COSM436924 rs1401664456 |
864 | R>C | breast [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs747587135 | 867 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs755285892 | 867 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs755285892 | 867 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1443856811 | 868 | M>I | No | gnomAD | |
| rs908878176 | 868 | M>V | No | TOPMed | |
| rs748285616 | 869 | E>K | No |
ExAC gnomAD |
|
| rs1373553825 | 870 | R>S | No |
TOPMed gnomAD |
|
| rs943021174 | 871 | V>F | No | TOPMed | |
| rs1597864729 | 871 | V>G | No | Ensembl | |
| rs1298881807 | 873 | P>L | No | gnomAD | |
| rs770017269 | 875 | A>T | No |
ExAC gnomAD |
|
| rs777901789 | 877 | D>Y | No |
ExAC gnomAD |
|
| rs1975134689 | 879 | E>D | No | TOPMed | |
| rs1480866105 | 881 | E>K | No |
TOPMed gnomAD |
|
| COSM981376 | 883 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs142384289 | 883 | E>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1451459534 | 884 | E>G | No | Ensembl | |
| rs1198013464 | 885 | G>D | No |
TOPMed gnomAD |
|
| rs1175645720 | 885 | G>S | No | gnomAD | |
| rs1324654826 | 888 | V>I | No | gnomAD | |
| rs1975135302 | 889 | P>L | No |
TOPMed gnomAD |
|
| rs779006452 | 889 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1361813822 | 893 | P>H | No | gnomAD | |
| rs1361813822 | 893 | P>R | No | gnomAD | |
| rs1262894595 | 894 | E>K | No |
TOPMed gnomAD |
|
| rs761602629 | 898 | A>S | No |
ExAC gnomAD |
|
| rs761602629 | 898 | A>T | No |
ExAC gnomAD |
|
| rs772986886 | 899 | A>V | No |
ExAC gnomAD |
|
| rs762745722 | 900 | E>G | No |
ExAC gnomAD |
|
| rs1389558261 | 901 | E>A | No | TOPMed | |
| COSM3519480 | 904 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1975136198 | 905 | S>N | No | Ensembl | |
| rs758990694 | 907 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs199975737 | 907 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs199975737 | 907 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs2143392523 RCV001532296 |
908 | M>I | No |
ClinVar Ensembl dbSNP |
|
| rs78970955 | 909 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs78970955 | 909 | P>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 909 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs867007726 | 911 | A>S | No | Ensembl | |
| rs147062279 | 912 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1348992399 | 913 | P>L | No | gnomAD | |
| rs368398299 | 913 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs779132007 | 914 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1246977283 | 915 | S>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3519482 rs549971892 |
915 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1597865583 | 915 | S>P | No | Ensembl | |
| TCGA novel | 915 | S>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748096408 | 916 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs200436795 | 917 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs773884974 | 919 | S>A | No |
ExAC gnomAD |
|
| rs1332298867 | 919 | S>L | No | TOPMed | |
| rs1975138165 | 920 | S>N | No | TOPMed | |
| rs1158891119 | 921 | W>C | No |
TOPMed gnomAD |
|
| rs138073126 | 923 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs138073126 | 923 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs147017530 | 923 | R>W | No |
ESP ExAC gnomAD |
|
| rs764619422 | 924 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs757633145 | 925 | S>A | No |
ExAC gnomAD |
|
| rs1975138827 | 925 | S>L | No | Ensembl | |
| rs200925951 | 926 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs765376957 | 926 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs1975138917 RCV001268850 |
927 | L>missing | No |
ClinVar dbSNP |
|
| rs748090224 | 928 | M>V | No |
ExAC gnomAD |
|
| rs1487182909 | 929 | E>Q | No | Ensembl | |
| rs756073486 | 931 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs756073486 | 931 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1463063506 | 931 | D>N | No | TOPMed | |
| rs756073486 | 931 | D>V | No |
ExAC TOPMed gnomAD |
|
|
COSM3519483 rs777872350 |
932 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs774048138 | 935 | R>C | No |
ExAC gnomAD |
|
| rs1248020072 | 935 | R>H | No | gnomAD | |
| rs745563559 | 936 | R>C | No |
ExAC gnomAD |
|
| rs1352623558 | 936 | R>H | No |
TOPMed gnomAD |
|
| rs761947051 | 937 | G>S | No | Ensembl | |
|
rs1182319112 COSM3519484 |
938 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1037994696 | 938 | R>H | No |
TOPMed gnomAD |
|
| rs1037994696 | 938 | R>P | No |
TOPMed gnomAD |
|
| rs771587184 | 940 | R>C | No |
ExAC gnomAD |
|
| rs1470355699 | 943 | K>R | No | gnomAD | |
| rs1975141248 | 944 | Q>* | No | TOPMed | |
| TCGA novel | 944 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1975141402 | 945 | E>D | No | gnomAD | |
| rs1282468836 | 945 | E>K | No | TOPMed | |
| rs777127538 | 948 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs764527829 | 948 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1285766718 | 952 | L>M | No |
TOPMed gnomAD |
|
| rs2143393187 | 952 | L>P | No | Ensembl | |
| rs1226113136 | 953 | Q>* | No | gnomAD | |
| rs1975142405 | 954 | G>C | No | Ensembl | |
| rs1030067320 | 954 | G>D | No |
TOPMed gnomAD |
|
| rs1196017111 | 955 | S>A | No | gnomAD | |
| rs777782427 | 958 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs200230098 | 958 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA gnomAD |
| rs1477435318 | 959 | G>C | No | gnomAD | |
| rs757104315 | 960 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs1168718880 | 961 | G>A | No | gnomAD | |
| COSM1384501 | 962 | L>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777114824 | 963 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1382144063 | 964 | R>S | No | gnomAD | |
| rs1213139314 | 966 | P>L | No |
TOPMed gnomAD |
|
| rs1975144436 | 966 | P>S | No | TOPMed | |
| rs1292911568 | 967 | A>T | No | gnomAD | |
| rs773411038 | 968 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs763396080 | 968 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1975144987 | 969 | F>L | No | TOPMed | |
| rs766450219 | 970 | V>M | No |
ExAC TOPMed gnomAD |
|
|
rs202059928 RCV001723330 |
971 | P>S | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs202059928 | 971 | P>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767762952 | 973 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1975145327 | 974 | D>E | No | TOPMed | |
| rs550136320 | 974 | D>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1398332598 | 975 | C>S | No |
TOPMed gnomAD |
|
| rs1184702686 | 975 | C>Y | No | gnomAD | |
| rs1340165720 | 977 | L>P | No | TOPMed | |
| rs1304103979 | 978 | R>C | No | TOPMed | |
| rs1975145527 | 978 | R>H | No | TOPMed | |
| rs867942178 | 980 | P>S | No | gnomAD | |
| rs1348637296 | 982 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 985 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs556299467 | 985 | P>S | No |
1000Genomes gnomAD |
|
| rs1975145882 | 986 | Q>* | No | TOPMed | |
| rs774122476 | 986 | Q>R | No | gnomAD | |
| rs1975146060 | 987 | H>Y | No | TOPMed | |
| rs193144796 | 988 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs376256028 | 988 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs747689834 | 989 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1382332110 | 990 | S>C | No | gnomAD | |
| rs1382332110 | 990 | S>F | No | gnomAD | |
| rs367650984 | 991 | W>C | No |
ESP TOPMed gnomAD |
|
| rs770147759 | 991 | W>R | No |
ExAC gnomAD |
|
| rs1264644495 | 993 | G>E | No |
TOPMed gnomAD |
|
| rs1192600230 | 993 | G>R | No | Ensembl | |
| rs773779354 | 994 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1975146555 | 995 | G>R | No | Ensembl | |
| rs1975146828 | 998 | E>G | No | Ensembl | |
| rs759886036 | 999 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs143987985 | 1002 | P>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs541004226 | 1003 | L>I | No |
1000Genomes ExAC gnomAD |
|
| rs2143393964 | 1003 | L>P | No | Ensembl | |
| rs370095783 | 1005 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1946495998 | 1005 | P>H | No | gnomAD | |
| rs370095783 | 1005 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1006 | P>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1157330216 | 1008 | E>V | No | gnomAD | |
| rs373574368 | 1010 | T>A | No |
ESP ExAC gnomAD |
|
| rs747596325 | 1010 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs747596325 | 1010 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs778134222 | 1012 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs755651054 | 1012 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1403257098 | 1012 | H>Y | No | gnomAD | |
| rs749860382 | 1013 | P>L | No |
ExAC gnomAD |
|
| rs1975148052 | 1014 | A>T | No |
TOPMed gnomAD |
|
| rs375145620 | 1015 | T>I | No |
ESP TOPMed gnomAD |
|
| rs1014528756 | 1015 | T>P | No | Ensembl | |
| rs1317509272 | 1016 | P>L | No | gnomAD | |
| rs1308853602 | 1016 | P>S | No | gnomAD | |
| rs185185243 | 1017 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs772370282 | 1018 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs775861610 | 1018 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs764293529 | 1019 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs760667669 | 1019 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1975148935 | 1020 | P>R | No | TOPMed | |
| rs369620316 | 1021 | P>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs369620316 | 1021 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1975149290 | 1022 | S>N | No | gnomAD | |
| rs1310634391 | 1023 | P>L | No | TOPMed | |
| rs1975149350 | 1023 | P>S | No | gnomAD | |
| rs141225452 | 1024 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs767087719 | 1025 | R>S | No |
ExAC gnomAD |
|
| rs1975149816 | 1026 | S>Y | No | Ensembl | |
| rs1299738298 | 1028 | H>R | No | gnomAD | |
| rs1439556561 | 1028 | H>Y | No | gnomAD | |
| rs755562244 | 1029 | P>L | No |
ExAC TOPMed gnomAD |
|
|
COSM3193368 rs139120839 |
1030 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs139120839 | 1030 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs62072492 | 1030 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1975150804 | 1036 | G>E | No | TOPMed | |
| rs1838334213 | 1038 | G>D | No |
TOPMed gnomAD |
|
| rs1159586653 | 1038 | G>S | No |
TOPMed gnomAD |
|
| rs775842918 | 1039 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs200388087 | 1041 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1202209298 | 1042 | G>A | No |
TOPMed gnomAD |
|
| rs1202209298 | 1042 | G>E | No |
TOPMed gnomAD |
|
| rs1202209298 | 1042 | G>V | No |
TOPMed gnomAD |
|
| rs1975151903 | 1045 | S>P | No | TOPMed | |
| TCGA novel | 1046 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767353539 | 1047 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs1407648265 | 1047 | Q>H | No | gnomAD | |
| rs767353539 | 1047 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1172108746 | 1048 | P>S | No | gnomAD | |
| rs1975152253 | 1049 | E>K | No | Ensembl | |
| rs570522657 | 1050 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1404147812 | 1050 | P>T | No | gnomAD | |
| rs1975152401 | 1051 | Q>P | No |
TOPMed gnomAD |
|
| rs760414992 | 1052 | H>P | No |
ExAC gnomAD |
|
| rs760624493 | 1053 | F>* | No |
ExAC gnomAD |
|
| rs1338826212 | 1053 | F>L | No | gnomAD | |
| rs763684173 | 1055 | P>L | No |
ExAC gnomAD |
|
| COSM981382 | 1055 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1163172391 | 1057 | K>R | No | gnomAD | |
| rs1975152813 | 1059 | N>D | No |
TOPMed gnomAD |
|
| rs1567729811 | 1059 | N>S | No | Ensembl | |
| rs1454815005 | 1060 | S>F | No |
TOPMed gnomAD |
|
| rs753338117 | 1060 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs1454815005 | 1060 | S>Y | No |
TOPMed gnomAD |
|
| rs1401065423 | 1061 | Y>* | No |
TOPMed gnomAD |
|
| rs756839157 | 1061 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs756839157 | 1061 | Y>N | No |
ExAC TOPMed gnomAD |
|
| rs751031419 | 1063 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1200208611 | 1065 | P>S | No | gnomAD | |
| rs1200208611 | 1065 | P>T | No | gnomAD | |
| rs1975153455 | 1066 | Q>P | No | Ensembl | |
| rs780508973 | 1067 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1474155917 | 1067 | P>S | No | gnomAD | |
| rs1474155917 | 1067 | P>T | No | gnomAD | |
| rs1975153744 | 1068 | Y>* | No | gnomAD | |
| rs1975153692 | 1068 | Y>D | No | Ensembl | |
| rs200907250 | 1069 | P>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs921895034 | 1069 | P>Q | No |
TOPMed gnomAD |
|
| rs200907250 | 1069 | P>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1367200582 | 1070 | A>D | No | gnomAD | |
| rs781287137 | 1071 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs781287137 | 1071 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1324564327 | 1072 | R>L | No |
TOPMed gnomAD |
|
| rs1324564327 | 1072 | R>Q | No |
TOPMed gnomAD |
|
|
COSM981383 rs748194713 |
1072 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1299876878 | 1073 | P>H | No |
TOPMed gnomAD |
|
| rs1299876878 | 1073 | P>L | No |
TOPMed gnomAD |
|
| rs1245516424 | 1075 | G>A | No |
TOPMed gnomAD |
|
| rs1245516424 | 1075 | G>E | No |
TOPMed gnomAD |
|
| rs1245516424 | 1075 | G>V | No |
TOPMed gnomAD |
|
| rs199800684 | 1077 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs759515240 | 1077 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs759515240 | 1077 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs759515240 | 1077 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1597866385 | 1078 | Y>F | No | Ensembl | |
| rs1284373251 | 1078 | Y>H | No | Ensembl | |
| rs1597866385 | 1078 | Y>S | No | Ensembl | |
| rs1360818803 | 1079 | P>T | No | TOPMed | |
| rs753993722 | 1080 | P>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763743652 | 1080 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs763743652 | 1080 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs763743652 | 1080 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs760407824 | 1080 | P>S | No |
ExAC gnomAD |
|
| rs764889837 | 1081 | Y>F | No |
ExAC gnomAD |
|
| rs761397495 | 1081 | Y>H | No |
ExAC gnomAD |
|
| TCGA novel | 1081 | Y>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs146157309 | 1082 | T>A | No |
ESP TOPMed gnomAD |
|
| rs1157739071 | 1083 | T>I | No |
TOPMed gnomAD |
|
| rs1382748594 | 1084 | P>A | No |
TOPMed gnomAD |
|
| COSM3519491 | 1084 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs570367142 | 1085 | P>L | No |
1000Genomes TOPMed gnomAD |
|
| rs570367142 | 1085 | P>Q | No |
1000Genomes TOPMed gnomAD |
|
| rs570367142 | 1085 | P>R | No |
1000Genomes TOPMed gnomAD |
|
| rs1975156625 | 1085 | P>S | No | TOPMed | |
| rs750930666 | 1086 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs139942468 | 1086 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1975157135 | 1087 | M>I | No | TOPMed | |
| rs1975157061 | 1087 | M>T | No | TOPMed | |
| rs1313221020 | 1088 | R>K | No | gnomAD | |
| rs535655372 | 1089 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs535655372 | 1089 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs780451511 | 1089 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1161457987 | 1090 | Q>R | No | TOPMed | |
| rs755305909 | 1091 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs201800868 | 1091 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201800868 | 1091 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs755305909 | 1091 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1225965913 | 1092 | S>F | No | gnomAD | |
| rs376866611 | 1093 | A>V | No |
ESP TOPMed gnomAD |
|
| rs867276857 | 1095 | D>N | No | gnomAD | |
| rs866339516 | 1097 | K>E | No |
TOPMed gnomAD |
|
| COSM3519492 | 1097 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs866339516 | 1097 | K>Q | No |
TOPMed gnomAD |
|
| rs749401324 | 1097 | K>R | No |
ExAC gnomAD |
|
| rs368230467 | 1098 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1597866526 | 1099 | S>N | No | Ensembl | |
| rs1471002106 | 1099 | S>R | No |
TOPMed gnomAD |
|
| rs999898090 | 1100 | G>V | No |
TOPMed gnomAD |
|
| rs1469384742 | 1102 | A>T | No | gnomAD | |
| rs775301108 | 1103 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1404684457 | 1104 | V>S | No |
TOPMed gnomAD |
1 associated diseases with O43896
[MIM: 611302]: Spastic ataxia 2, autosomal recessive (SPAX2)
A neurologic disorder characterized by cerebellar ataxia, dysarthria, and variable spasticity of the lower limbs. Cognition is not affected. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A neurologic disorder characterized by cerebellar ataxia, dysarthria, and variable spasticity of the lower limbs. Cognition is not affected. . Note=The disease is caused by variants affecting the gene represented in this entry.
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| axon cytoplasm | Any cytoplasm that is part of a axon. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| kinesin complex | Any complex that includes a dimer of molecules from the kinesin superfamily, a group of related proteins that contain an extended region of predicted alpha-helical coiled coil in the main chain that likely produces dimerization. The native complexes of several kinesin family members have also been shown to contain additional peptides, often designated light chains as all of the noncatalytic subunits that are currently known are smaller than the chain that contains the motor unit. Kinesin complexes generally possess a force-generating enzymatic activity, or motor, which converts the free energy of the gamma phosphate bond of ATP into mechanical work. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| cytoskeletal motor activity | Generation of force resulting in movement, for example along a microfilament or microtubule, or in torque resulting in membrane scission or rotation of a flagellum. The energy required is obtained either from the hydrolysis of a nucleoside triphosphate or by an electrochemical proton gradient (proton-motive force). |
| microtubule binding | Binding to a microtubule, a filament composed of tubulin monomers. |
| plus-end-directed microtubule motor activity | A motor activity that generates movement along a microtubule toward the plus end, driven by ATP hydrolysis. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| anterograde neuronal dense core vesicle transport | The directed movement of substances in neuronal dense core vesicles along axonal microtubules towards the presynapse. |
| cytoskeleton-dependent intracellular transport | The directed movement of substances along cytoskeletal fibers such as microfilaments or microtubules within a cell. |
| microtubule-based movement | A microtubule-based process that results in the movement of organelles, other microtubules, or other cellular components. Examples include motor-driven movement along microtubules and movement driven by polymerization or depolymerization of microtubules. |
| retrograde neuronal dense core vesicle transport | The directed movement of neuronal dense core vesicles along axonal microtubules towards the cell body. |
| retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | The directed movement of substances from the Golgi back to the endoplasmic reticulum, mediated by vesicles bearing specific protein coats such as COPI or COG. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P33176 | KIF5B | Kinesin-1 heavy chain | Homo sapiens (Human) | EV |
| Q2M1P5 | KIF7 | Kinesin-like protein KIF7 | Homo sapiens (Human) | EV |
| Q12840 | KIF5A | Kinesin heavy chain isoform 5A | Homo sapiens (Human) | EV |
| O60282 | KIF5C | Kinesin heavy chain isoform 5C | Homo sapiens (Human) | EV |
| Q86VH2 | KIF27 | Kinesin-like protein KIF27 | Homo sapiens (Human) | SS |
| Q7Z4S6 | KIF21A | Kinesin-like protein KIF21A | Homo sapiens (Human) | EV |
| O75037 | KIF21B | Kinesin-like protein KIF21B | Homo sapiens (Human) | EV |
| Q9P2E2 | KIF17 | Kinesin-like protein KIF17 | Homo sapiens (Human) | EV |
| O00139 | KIF2A | Kinesin-like protein KIF2A | Homo sapiens (Human) | PR |
| Q9NQT8 | KIF13B | Kinesin-like protein KIF13B | Homo sapiens (Human) | EV |
| Q9H1H9 | KIF13A | Kinesin-like protein KIF13A | Homo sapiens (Human) | SS |
| G5EFQ4 | klp-6 | Kinesin-like protein | Caenorhabditis elegans | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGASVKVAV | RVRPFNARET | SQDAKCVVSM | QGNTTSIINP | KQSKDAPKSF | TFDYSYWSHT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| STEDPQFASQ | QQVYRDIGEE | MLLHAFEGYN | VCIFAYGQTG | AGKSYTMMGR | QEPGQQGIVP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QLCEDLFSRV | SENQSAQLSY | SVEVSYMEIY | CERVRDLLNP | KSRGSLRVRE | HPILGPYVQD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LSKLAVTSYA | DIADLMDCGN | KARTVAATNM | NETSSRSHAV | FTIVFTQRCH | DQLTGLDSEK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VSKISLVDLA | GSERADSSGA | RGMRLKEGAN | INKSLTTLGK | VISALADMQS | KKRKSDFIPY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RDSVLTWLLK | ENLGGNSRTA | MIAALSPADI | NYEETLSTLR | YADRTKQIRC | NAIINEDPNA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RLIRELQEEV | ARLRELLMAQ | GLSASALEGL | KTEEGSVRGA | LPAVSSPPAP | VSPSSPTTHN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GELEPSFSPN | TESQIGPEEA | MERLQETEKI | IAELNETWEE | KLRKTEALRM | EREALLAEMG |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VAVREDGGTV | GVFSPKKTPH | LVNLNEDPLM | SECLLYHIKD | GVTRVGQVDM | DIKLTGQFIR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EQHCLFRSIP | QPDGEVVVTL | EPCEGAETYV | NGKLVTEPLV | LKSGNRIVMG | KNHVFRFNHP |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EQARLERERG | VPPPPGPPSE | PVDWNFAQKE | LLEQQGIDIK | LEMEKRLQDL | ENQYRKEKEE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| ADLLLEQQRL | YADSDSGDDS | DKRSCEESWR | LISSLREQLP | PTTVQTIVKR | CGLPSSGKRR |
| 730 | 740 | 750 | 760 | 770 | 780 |
| APRRVYQIPQ | RRRLQGKDPR | WATMADLKMQ | AVKEICYEVA | LADFRHGRAE | IEALAALKMR |
| 790 | 800 | 810 | 820 | 830 | 840 |
| ELCRTYGKPD | GPGDAWRAVA | RDVWDTVGEE | EGGGAGSGGG | SEEGARGAEV | EDLRAHIDKL |
| 850 | 860 | 870 | 880 | 890 | 900 |
| TGILQEVKLQ | NSSKDRELQA | LRDRMLRMER | VIPLAQDHED | ENEEGGEVPW | APPEGSEAAE |
| 910 | 920 | 930 | 940 | 950 | 960 |
| EAAPSDRMPS | ARPPSPPLSS | WERVSRLMEE | DPAFRRGRLR | WLKQEQLRLQ | GLQGSGGRGG |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| GLRRPPARFV | PPHDCKLRFP | FKSNPQHRES | WPGMGSGEAP | TPLQPPEEVT | PHPATPARRP |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| PSPRRSHHPR | RNSLDGGGRS | RGAGSAQPEP | QHFQPKKHNS | YPQPPQPYPA | QRPPGPRYPP |
| 1090 | 1100 | ||||
| YTTPPRMRRQ | RSAPDLKESG | AAV |