O43739
Gene name |
CYTH3 |
Protein name |
Cytohesin-3 |
Names |
ARF nucleotide-binding site opener 3, Protein ARNO3, General receptor of phosphoinositides 1, Grp1, PH, SEC7 and coiled-coil domain-containing protein 3 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9265 |
EC number |
|
Protein Class |
GUANYL-NUCLEOTIDE EXCHANGE FACTOR (PTHR10663) |
Descriptions
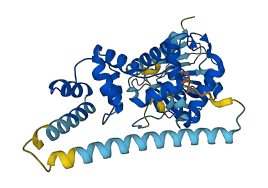
Cytohesin-3 (CYTH3) promotes guanine-nucleotide exchange on Arf1 and Arf6. Cytohesins share a modular architecture consisting of heptad repeats, a Sec7 domain with exchange activity for Arf1 and Arf6, a PH domain that binds phosphatidylinositol (PI) polyphosphates, and a C-terminal helix (CtH) that overlaps with a polybasic region (PBR). Cytohesins are autoinhibited by the Sec7-PH linker (252-264) and CtH/PBR (380-399), which obstruct substrate binding. The autoinhibition can be relieved by Arf6-GTP binding in the presence of the PIP3 head group.
Autoinhibitory domains (AIDs)
Target domain |
63-251 (Sec7 domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Structural analysis |
Target domain |
63-251 (Sec7 domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure
4 structures for O43739
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4KAX | X-ray | 185 A | B | 247-400 | PDB |
| 6U3E | EM | 5300 A | A/B | 13-400 | PDB |
| 6U3G | EM | 5300 A | A/B | 13-400 | PDB |
| AF-O43739-F1 | Predicted | AlphaFoldDB |
232 variants for O43739
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs763559891 CA4153312 |
2 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs1420755972 CA366762009 |
8 | E>G | No |
ClinGen TOPMed |
|
|
rs775683144 CA4153311 |
8 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA366761989 rs1473136310 |
9 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
CA366762001 rs1158582089 |
9 | G>S | No |
ClinGen gnomAD |
|
|
rs1185635429 CA366761949 |
11 | G>A | No |
ClinGen gnomAD |
|
|
rs1185635429 CA366761944 |
11 | G>D | No |
ClinGen gnomAD |
|
|
CA366761961 rs1370295973 |
11 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1333318616 CA366765886 |
13 | P>A | No |
ClinGen gnomAD |
|
|
rs1322216840 CA366765876 |
13 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA4153298 rs757041462 |
18 | L>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366765627 rs11978706 |
25 | L>I | No |
ClinGen TOPMed gnomAD |
|
|
CA366765624 rs11978706 |
25 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA366765584 rs1478926594 |
26 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1478926594 CA366765586 |
26 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA366765591 rs1172934202 |
26 | D>G | No |
ClinGen gnomAD |
|
|
rs1423244306 CA366765602 |
26 | D>H | No |
ClinGen gnomAD |
|
|
rs142160586 CA4153297 |
28 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs757875265 CA4153295 |
29 | R>* | No |
ClinGen ExAC gnomAD |
|
|
rs375159013 CA4153294 |
29 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA366765536 rs1278961525 |
30 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
rs535847871 CA4153293 |
31 | K>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
COSM1451750 CA366765480 rs1273526452 |
32 | K>N | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA366765497 rs1347583472 |
32 | K>Q | No |
ClinGen gnomAD |
|
|
rs1234230533 CA366765468 |
33 | E>K | No |
ClinGen gnomAD |
|
|
rs759828701 CA4153292 |
35 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366765422 rs1333421745 |
36 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA366765427 rs1200247005 |
36 | D>N | No |
ClinGen TOPMed |
|
|
rs1335112339 CA366765402 |
37 | D>E | No |
ClinGen gnomAD |
|
|
CA366765414 rs988520269 |
37 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
CA4153291 rs754267657 |
37 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs988520269 CA153291219 |
37 | D>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA366765395 rs958095503 |
38 | I>N | No |
ClinGen TOPMed gnomAD |
|
|
rs958095503 CA153291208 |
38 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
rs568410526 CA4153290 |
38 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA4153256 rs200739052 |
41 | L>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1583761414 CA366763806 |
41 | L>P | No |
ClinGen Ensembl |
|
|
CA4153255 rs759582996 |
42 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4153254 rs776737393 |
42 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366763757 rs1377228364 |
43 | Y>C | No |
ClinGen gnomAD |
|
|
rs942417299 CA153289466 |
43 | Y>H | No |
ClinGen Ensembl |
|
|
rs1421072467 CA366763716 |
44 | E>D | No |
ClinGen gnomAD |
|
|
rs777415508 CA4153251 |
48 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA4153249 rs747679549 |
49 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA4153250 rs771686875 |
49 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA4153246 rs754304399 |
53 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA4153247 rs755446683 |
53 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780562697 CA4153245 |
54 | N>D | No |
ClinGen ExAC TOPMed |
|
|
CA4153244 rs756445114 |
54 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1355494506 CA366763444 |
56 | T>I | No |
ClinGen TOPMed |
|
|
rs142012512 CA4153241 |
58 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs142012512 CA366763406 |
58 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4153240 rs751671861 |
60 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs1245879097 CA366763204 |
61 | S>R | No |
ClinGen TOPMed |
|
|
rs1204083467 CA366763323 |
61 | S>T | No |
ClinGen TOPMed |
|
|
rs761618708 CA153289133 |
62 | K>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4153213 rs761618708 |
62 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs184752712 CA4153212 COSM1451748 |
63 | T>M | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
CA4153210 rs748781445 |
65 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs1360136315 CA366763107 |
66 | R>M | No |
ClinGen gnomAD |
|
|
rs770301160 CA4153208 |
66 | R>S | No |
ClinGen ExAC gnomAD |
|
|
rs746407066 COSM216788 CA4153207 |
67 | N>S | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA366762995 rs1157347949 |
70 | I>M | No |
ClinGen TOPMed |
|
|
CA4153205 rs781740165 |
74 | R>S | No |
ClinGen ExAC gnomAD |
|
|
rs757500800 CA4153204 |
76 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs747394665 CA4153203 |
78 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs1257506365 CA366762682 |
79 | M>I | No |
ClinGen gnomAD |
|
|
rs1483430778 CA366762708 |
79 | M>T | No |
ClinGen gnomAD |
|
|
CA4153201 rs758761652 |
81 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs61753121 CA4153199 |
82 | K>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4153178 rs757249158 |
85 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs748143048 CA153284882 |
89 | I>T | No |
ClinGen Ensembl |
|
|
rs763922164 CA4153176 |
89 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1163074197 CA366759043 |
90 | E>V | No |
ClinGen gnomAD |
|
|
CA366759036 rs1474732204 |
91 | N>D | No |
ClinGen gnomAD |
|
|
CA153284875 rs1003936954 |
92 | D>E | No |
ClinGen Ensembl |
|
|
rs1562880430 CA366758985 |
95 | Q>* | No |
ClinGen Ensembl |
|
|
rs1445189514 CA366758975 |
95 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA4153175 rs762754518 |
96 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs1357854504 CA366758928 |
99 | E>G | No |
ClinGen gnomAD |
|
|
rs555480535 CA4153172 |
101 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA366758871 rs761315277 |
103 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366758883 rs1279208163 |
103 | Q>K | No |
ClinGen gnomAD |
|
|
rs372746038 CA4153170 |
103 | Q>R | No |
ClinGen ESP ExAC gnomAD |
|
|
CA366758857 rs1318248443 |
104 | F>L | No |
ClinGen gnomAD |
|
|
CA153284844 rs886793147 |
107 | K>R | No |
ClinGen TOPMed |
|
|
CA153284842 rs1056104354 |
110 | G>V | No |
ClinGen Ensembl |
|
|
CA366758776 rs1369164904 |
112 | N>S | No |
ClinGen TOPMed |
|
|
rs748550130 CA4153166 |
115 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs768979781 CA4153164 |
116 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA4153165 rs780958252 |
116 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4153160 rs375065839 |
118 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4153161 rs757161085 |
118 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs780144942 CA366758716 |
118 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs780144942 CA4153162 |
118 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA366758692 rs1227061961 |
119 | Y>C | No |
ClinGen TOPMed |
|
|
rs750011072 CA4153131 |
124 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA366757836 rs1381663136 |
125 | E>* | No |
ClinGen gnomAD |
|
|
rs768133140 CA4153130 |
127 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA366757794 rs1469291191 |
129 | K>E | No |
ClinGen gnomAD |
|
|
CA4153129 rs762421588 |
129 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA4153127 rs764467386 |
133 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA366757672 rs1355095333 |
139 | E>G | No |
ClinGen gnomAD |
|
|
rs939848869 CA153281285 |
144 | N>S | No |
ClinGen gnomAD |
|
|
CA4153124 rs770063156 |
147 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA366757571 rs1468695404 |
147 | Q>P | No |
ClinGen TOPMed |
|
|
rs766524130 CA4153083 |
153 | L>* | No |
ClinGen ExAC gnomAD |
|
|
rs1161617589 CA366756493 |
155 | S>T | No |
ClinGen gnomAD |
|
|
rs776236256 CA153279414 |
156 | F>V | No |
ClinGen Ensembl |
|
|
rs768605779 CA4153080 |
174 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA4153078 rs775587767 |
176 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs1212146089 CA366756336 |
177 | C>Y | No |
ClinGen gnomAD |
|
|
rs368016139 CA4153077 |
178 | L>V | No |
ClinGen 1000Genomes ESP ExAC gnomAD |
|
|
rs770560431 CA4153074 |
179 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs746501936 CA4153073 |
181 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1381672202 CA366756280 |
181 | P>S | No |
ClinGen TOPMed |
|
|
CA366756271 rs1347589526 |
182 | G>R | No |
ClinGen TOPMed |
|
|
rs993781059 CA153279345 |
185 | Q>* | No |
ClinGen Ensembl |
|
|
rs1562876400 COSM1731460 CA366756201 |
187 | T>A | liver [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA366756059 rs1349682736 |
189 | T>K | No |
ClinGen gnomAD |
|
|
CA153279176 rs763037020 |
192 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs763037020 CA4153047 |
192 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs1346075897 CA366755932 |
195 | F>V | No |
ClinGen gnomAD |
|
|
rs750567643 CA153279170 |
197 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs950534428 CA153279171 |
197 | I>V | No |
ClinGen TOPMed |
|
|
rs1332923883 CA366755840 |
199 | M>L | No |
ClinGen gnomAD |
|
|
CA366755773 rs1409635362 |
202 | T>P | No |
ClinGen gnomAD |
|
|
CA4153044 rs767853400 |
204 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1480473078 CA366755717 |
205 | H>D | No |
ClinGen TOPMed |
|
|
CA366755671 rs1404386443 |
208 | N>S | No |
ClinGen gnomAD |
|
|
rs1411654026 CA366755661 |
209 | V>L | No |
ClinGen gnomAD |
|
|
rs1189156962 CA366755647 |
210 | R>C | No |
ClinGen gnomAD |
|
|
rs771188198 CA153279166 |
211 | D>E | No |
ClinGen Ensembl |
|
|
rs1388717285 CA366755546 |
214 | T>M | No |
ClinGen gnomAD |
|
|
CA4153041 rs150471410 |
215 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA366755531 rs1163001665 |
216 | E>A | No |
ClinGen TOPMed |
|
|
rs759445103 CA366755496 |
218 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs776655596 CA4153039 |
219 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA153279136 COSM3784080 rs749850896 |
220 | A>T | prostate [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA366755426 rs1168514895 |
223 | R>C | No |
ClinGen TOPMed |
|
|
CA366755423 rs1293661319 |
223 | R>H | No |
ClinGen gnomAD |
|
|
rs1283538829 CA366755406 |
224 | G>D | No |
ClinGen gnomAD |
|
|
rs1353408890 CA366755411 |
224 | G>S | No |
ClinGen gnomAD |
|
|
rs772795279 CA4153036 |
227 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1562876126 CA366755365 |
227 | E>K | No |
ClinGen Ensembl |
|
|
rs1353736050 CA366755345 |
228 | G>C | No |
ClinGen TOPMed |
|
|
CA4153034 rs141576733 |
229 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs773890198 CA4153033 |
231 | L>F | No |
ClinGen ExAC |
|
|
CA366755279 rs1562876096 |
232 | P>S | No |
ClinGen Ensembl |
|
|
rs1338673965 CA366755259 |
233 | E>G | No |
ClinGen TOPMed |
|
|
CA4153031 rs749707136 |
234 | E>* | No |
ClinGen ExAC gnomAD |
|
|
rs780641749 CA4153030 |
234 | E>V | No |
ClinGen ExAC gnomAD |
|
|
CA366755238 rs1416261935 |
235 | L>V | No |
ClinGen gnomAD |
|
|
rs369539860 CA4152997 |
238 | N>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4152996 rs530427375 |
238 | N>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA153278754 rs753297433 |
245 | N>S | No |
ClinGen Ensembl |
|
|
rs905373667 CA153278748 |
252 | E>Q | No |
ClinGen TOPMed |
|
|
rs777312110 CA4152992 |
254 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA366755069 rs1306610408 |
257 | D>N | No |
ClinGen gnomAD |
|
|
CA366755001 rs1252407636 |
264 | N>D | No |
ClinGen TOPMed |
|
|
rs748146676 CA366754987 |
264 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1160139638 CA366754992 |
264 | N>S | No |
ClinGen gnomAD |
|
|
CA366754967 rs1181253011 |
266 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1181253011 CA366754962 |
266 | D>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA4152983 rs781110781 |
267 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA366754809 rs1583733752 |
274 | L>R | No |
ClinGen Ensembl |
|
|
CA366754799 rs1314816675 |
275 | G>C | No |
ClinGen gnomAD |
|
|
rs1297011969 CA366753145 |
278 | R>C | No |
ClinGen gnomAD |
|
|
CA4152937 rs750874462 |
278 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA153274950 rs1038623799 |
280 | K>R | No |
ClinGen TOPMed |
|
|
rs762223583 CA4152934 |
284 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs751931620 CA4152933 |
284 | R>P | No |
ClinGen ExAC gnomAD |
|
|
rs1562872655 CA366753021 |
290 | T>I | No |
ClinGen Ensembl |
|
|
rs1461014386 CA366752890 |
299 | Y>H | No |
ClinGen gnomAD |
|
|
rs758752571 CA4152895 |
304 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366752643 rs1187006583 |
316 | I>T | No |
ClinGen gnomAD |
|
|
rs755203995 CA4152892 |
320 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA153274678 rs766331224 |
321 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754113042 CA4152891 |
321 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs1376956652 CA366752480 |
323 | R>Q | No |
ClinGen gnomAD |
|
|
CA4152889 rs760840529 |
323 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA366752446 rs1424853644 |
325 | P>S | No |
ClinGen gnomAD |
|
|
rs756444576 CA4152851 |
330 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA4152849 rs781368499 |
331 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
CA366752200 rs1365885009 |
331 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1410960157 CA366752177 |
332 | N>T | No |
ClinGen gnomAD |
|
|
CA4152846 rs563074951 |
333 | P>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA153274416 rs886677624 |
334 | S>R | No |
ClinGen Ensembl |
|
|
rs1429202543 CA366751978 |
343 | C>R | No |
ClinGen gnomAD |
|
|
rs551111893 CA4152845 |
345 | T>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs551111893 CA153274414 |
345 | T>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs551111893 CA4152844 |
345 | T>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA366751938 rs1408375758 |
346 | E>Q | No |
ClinGen gnomAD |
|
|
CA366751905 rs1182710124 |
348 | D>A | No |
ClinGen gnomAD |
|
|
rs1447043436 CA366751911 |
348 | D>H | No |
ClinGen TOPMed |
|
|
rs1447043436 CA366751913 |
348 | D>N | No |
ClinGen TOPMed |
|
|
CA4152839 rs772883077 |
349 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1483306780 CA366751866 |
350 | R>C | No |
ClinGen gnomAD |
|
|
CA366751853 rs1299916093 |
351 | V>M | No |
ClinGen TOPMed |
|
|
rs1583724229 CA366751821 |
352 | V>A | No |
ClinGen Ensembl |
|
|
rs1431386074 CA366751767 |
355 | N>H | No |
ClinGen gnomAD |
|
|
CA366751680 rs1301009923 |
358 | V>A | No |
ClinGen gnomAD |
|
|
rs749584345 CA153274369 |
359 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs948442724 CA153274349 |
360 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs371624575 COSM1091138 CA366751636 |
360 | R>L | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs371624575 CA4152833 |
360 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA366751645 rs948442724 |
360 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA366751552 rs746044558 |
364 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4152831 rs746044558 |
364 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4152832 rs746044558 |
364 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1409242756 CA366751569 |
364 | P>S | No |
ClinGen gnomAD |
|
|
rs867287441 CA366751520 |
365 | S>R | No |
ClinGen gnomAD |
|
|
CA4152825 rs539932644 |
366 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4152826 rs539932644 |
366 | P>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA366751502 rs539932644 |
366 | P>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs778687948 CA4152827 |
366 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs767216711 CA4152821 |
368 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA4152819 rs542536904 |
369 | K>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs144116589 CA4152814 |
376 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA572569582 rs1230704749 |
377 | K>N | No |
ClinGen gnomAD |
|
|
rs760964732 CA4152773 |
380 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1269852492 CA366750704 |
384 | P>T | No |
ClinGen gnomAD |
|
|
CA366750662 rs1332575110 |
386 | Y>F | No |
ClinGen gnomAD |
|
|
CA4152770 rs761847410 |
390 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs774460054 CA4152769 |
391 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs768742187 CA4152768 |
391 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4152766 rs780996917 |
394 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1432522202 CA366750521 |
395 | R>K | No |
ClinGen gnomAD |
|
|
CA153273884 rs965553144 |
397 | A>G | No |
ClinGen TOPMed |
|
|
rs1562871845 CA366750480 |
398 | N>H | No |
ClinGen Ensembl |
|
|
CA366750443 rs1197278095 |
400 | K>E | No |
ClinGen gnomAD |
|
|
CA4152765 rs770701820 |
400 | K>N | No |
ClinGen ExAC gnomAD |
No associated diseases with O43739
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR10663 | GUANYL-NUCLEOTIDE EXCHANGE FACTOR |
| PANTHER Subfamily | PTHR10663:SF320 | CYTOHESIN-3 |
| PANTHER Protein Class |
guanyl-nucleotide exchange factor
G-protein modulator protein-binding activity modulator |
|
| PANTHER Pathway Category | No pathway information available | |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| adherens junction | A cell-cell junction composed of the epithelial cadherin-catenin complex. The epithelial cadherins, or E-cadherins, of each interacting cell extend through the plasma membrane into the extracellular space and bind to each other. The E-cadherins bind to catenins on the cytoplasmic side of the membrane, where the E-cadherin-catenin complex binds to cytoskeletal components and regulatory and signaling molecules. |
| bicellular tight junction | An occluding cell-cell junction that is composed of a branching network of sealing strands that completely encircles the apical end of each cell in an epithelial sheet; the outer leaflets of the two interacting plasma membranes are seen to be tightly apposed where sealing strands are present. Each sealing strand is composed of a long row of transmembrane adhesion proteins embedded in each of the two interacting plasma membranes. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extrinsic component of cytoplasmic side of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to its cytoplasmic surface, but not integrated into the hydrophobic region. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| guanyl-nucleotide exchange factor activity | Stimulates the exchange of GDP to GTP on a signaling GTPase, changing its conformation to its active form. Guanine nucleotide exchange factors (GEFs) act by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP), which is more abundant in the cell under normal cellular physiological conditions. |
| phosphatidylinositol-3,4,5-trisphosphate binding | Binding to phosphatidylinositol-3,4,5-trisphosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 3', 4' and 5' positions. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| establishment of epithelial cell polarity | The specification and formation of anisotropic intracellular organization of an epithelial cell. |
| Golgi vesicle transport | The directed movement of substances into, out of or within the Golgi apparatus, mediated by vesicles. |
| positive regulation of cell adhesion | Any process that activates or increases the frequency, rate or extent of cell adhesion. |
| regulation of ARF protein signal transduction | Any process that modulates the frequency, rate or extent of ARF protein signal transduction. |
17 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KI41 | CYTH2 | Cytohesin-2 | Bos taurus (Bovine) | SS |
| Q9Y6D5 | ARFGEF2 | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | Homo sapiens (Human) | PR |
| Q9Y6D6 | ARFGEF1 | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 | Homo sapiens (Human) | PR |
| Q5JU85 | IQSEC2 | IQ motif and SEC7 domain-containing protein 2 | Homo sapiens (Human) | EV |
| Q9UPP2 | IQSEC3 | IQ motif and SEC7 domain-containing protein 3 | Homo sapiens (Human) | SS |
| Q6DN90 | IQSEC1 | IQ motif and SEC7 domain-containing protein 1 | Homo sapiens (Human) | EV |
| Q99418 | CYTH2 | Cytohesin-2 | Homo sapiens (Human) | SS |
| Q9UIA0 | CYTH4 | Cytohesin-4 | Homo sapiens (Human) | SS |
| Q15438 | CYTH1 | Cytohesin-1 | Homo sapiens (Human) | SS |
| Q92538 | GBF1 | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 | Homo sapiens (Human) | PR |
| P63034 | Cyth2 | Cytohesin-2 | Mus musculus (Mouse) | SS |
| Q80YW0 | Cyth4 | Cytohesin-4 | Mus musculus (Mouse) | SS |
| Q9QX11 | Cyth1 | Cytohesin-1 | Mus musculus (Mouse) | SS |
| O08967 | Cyth3 | Cytohesin-3 | Mus musculus (Mouse) | EV |
| P97694 | Cyth1 | Cytohesin-1 | Rattus norvegicus (Rat) | SS |
| P63035 | Cyth2 | Cytohesin-2 | Rattus norvegicus (Rat) | SS |
| P97696 | Cyth3 | Cytohesin-3 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDEDGGGEGG | GVPEDLSLEE | REELLDIRRR | KKELIDDIER | LKYEIAEVMT | EIDNLTSVEE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SKTTQRNKQI | AMGRKKFNMD | PKKGIQFLIE | NDLLQSSPED | VAQFLYKGEG | LNKTVIGDYL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GERDEFNIKV | LQAFVELHEF | ADLNLVQALR | QFLWSFRLPG | EAQKIDRMME | AFASRYCLCN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PGVFQSTDTC | YVLSFAIIML | NTSLHNHNVR | DKPTAERFIA | MNRGINEGGD | LPEELLRNLY |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ESIKNEPFKI | PEDDGNDLTH | TFFNPDREGW | LLKLGGGRVK | TWKRRWFILT | DNCLYYFEYT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TDKEPRGIIP | LENLSIREVE | DPRKPNCFEL | YNPSHKGQVI | KACKTEADGR | VVEGNHVVYR |
| 370 | 380 | 390 | |||
| ISAPSPEEKE | EWMKSIKASI | SRDPFYDMLA | TRKRRIANKK |