O43639
Gene name |
NCK2 (GRB4) |
Protein name |
Cytoplasmic protein NCK2 |
Names |
Growth factor receptor-bound protein 4, NCK adaptor protein 2, Nck-2, SH2/SH3 adaptor protein NCK-beta |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:8440 |
EC number |
|
Protein Class |
SH2-SH3 ADAPTOR PROTEIN-RELATED (PTHR19969) |
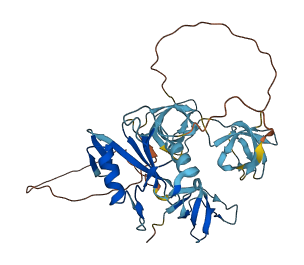
Descriptions
NCK2 is a cytoplasmic protein participating in the reorganization of the actin cytoskeleton and in the formation of the immunological synapses. The autoinhibitory domain locates at a conserved non-canonical (K/R)x(K/R)RxxS sequence, specifically at residues 60-110 linker between the SH3.1 and SH3.2. The autoinhibitory domain binds to the SH3.2 domain via acidic electrostatic potential on the SH3 domain surface and masks the proline-rich sequence (PRS) binding site of SH3.2 from its ligands. Only high affinity stable interactions can overcome the inhibitory interaction; such as phosphorylation of Ser85 at the interaction site.
Autoinhibitory domains (AIDs)
Target domain |
111-170 (SH3.2 domain) |
Relief mechanism |
PTM |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

10 structures for O43639
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1U5S | NMR | - | A | 192-262 | PDB |
| 1WX6 | NMR | - | A | 188-265 | PDB |
| 1Z3K | NMR | - | A | 283-380 | PDB |
| 2B86 | NMR | - | A | 1-59 | PDB |
| 2CIA | X-ray | 145 A | A | 284-380 | PDB |
| 2FRW | NMR | - | A | 114-170 | PDB |
| 2FRY | NMR | - | A | 198-256 | PDB |
| 2JXB | NMR | - | A | 1-59 | PDB |
| 4E6R | X-ray | 220 A | A/B | 114-170 | PDB |
| AF-O43639-F1 | Predicted | AlphaFoldDB |
284 variants for O43639
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA348103337 rs1195924667 |
4 | E>V | No |
ClinGen TOPMed |
|
|
rs201418674 CA1815020 |
6 | I>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs769339535 CA53754956 |
7 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769339535 CA1815022 |
7 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1573207897 CA348103358 |
8 | I>V | No |
ClinGen Ensembl |
|
|
rs774773148 CA1815023 |
10 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs768027105 CA1815025 |
15 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA348103423 rs1330679162 |
17 | Q>* | No |
ClinGen TOPMed |
|
|
rs750771445 CA1815026 |
23 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1050345687 CA53754988 |
26 | N>D | No |
ClinGen Ensembl |
|
|
CA348103507 rs1175209860 |
28 | R>Q | No |
ClinGen gnomAD |
|
|
rs1573208012 CA348103538 |
33 | D>G | No |
ClinGen Ensembl |
|
|
COSM1004780 COSM1004778 rs752670187 CA1815032 |
34 | D>N | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1371960767 CA348103561 |
36 | K>N | No |
ClinGen TOPMed |
|
|
rs765463213 CA53755019 |
37 | T>M | No |
ClinGen TOPMed |
|
|
rs368161463 CA1815036 |
40 | R>Q | No |
ClinGen ESP ExAC |
|
|
COSM1004781 CA1815035 rs746745967 |
40 | R>W | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed |
|
rs1573208133 CA348103594 |
41 | V>G | No |
ClinGen Ensembl |
|
|
rs1573208145 CA348103596 |
42 | R>G | No |
ClinGen Ensembl |
|
|
rs902978534 CA348103610 |
44 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
rs902978534 CA53755061 |
44 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA1815038 rs745514962 |
44 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA53755066 rs973694152 |
45 | A>V | No |
ClinGen TOPMed |
|
|
rs1282227137 CA348103635 |
48 | T>A | No |
ClinGen gnomAD |
|
|
rs775028062 CA1815040 |
48 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348103685 rs571195450 |
55 | Y>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs375461757 CA1815042 |
55 | Y>S | No |
ClinGen ESP ExAC gnomAD |
|
|
rs761188698 CA1815044 |
56 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs766753457 CA1815045 |
58 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
COSM1613254 CA348103700 rs1558871309 COSM1613253 |
58 | R>W | liver [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA348103736 rs1478926029 |
63 | K>R | No |
ClinGen gnomAD |
|
|
CA348103751 rs1395888799 |
65 | G>D | No |
ClinGen gnomAD |
|
|
CA348103766 rs1404717219 |
68 | V>M | No |
ClinGen gnomAD |
|
|
rs757054911 CA1815053 |
72 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA348103809 rs1220116381 |
74 | T>S | No |
ClinGen gnomAD |
|
|
CA348103839 rs751541163 |
77 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815071 rs751541163 |
77 | L>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751541163 CA53770544 |
77 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815072 rs757182579 |
78 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815073 rs767240597 |
80 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1468163686 CA348103863 |
81 | R>C | No |
ClinGen TOPMed |
|
|
rs1189638994 CA348103866 |
81 | R>L | No |
ClinGen gnomAD |
|
|
rs755727286 CA1815075 |
85 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA348103891 rs1162185719 |
85 | S>T | No |
ClinGen gnomAD |
|
|
COSM1564879 CA348103896 COSM1564878 rs1382749200 |
86 | A>T | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs913136866 COSM3772386 COSM3772387 CA53770563 |
86 | A>V | pancreas [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA348103900 rs1158424197 |
87 | R>G | No |
ClinGen gnomAD |
|
|
rs748935902 CA1815077 |
87 | R>P | No |
ClinGen ExAC gnomAD |
|
|
CA348103901 rs1158424197 |
87 | R>W | No |
ClinGen gnomAD |
|
|
rs1338072592 CA348103904 |
88 | D>N | No |
ClinGen gnomAD |
|
|
CA348103917 rs1382927137 |
89 | A>V | No |
ClinGen gnomAD |
|
|
rs973651345 CA53770568 |
90 | S>P | No |
ClinGen Ensembl |
|
|
rs747500810 CA1815080 |
92 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771417618 CA1815081 |
93 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1263830219 CA348103943 |
94 | S>T | No |
ClinGen TOPMed |
|
|
rs776872198 CA1815082 |
95 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201406196 CA53770597 |
96 | D>G | No |
ClinGen Ensembl |
|
|
rs770047813 CA1815084 |
96 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1360838761 CA348103963 |
97 | A>D | No |
ClinGen TOPMed |
|
|
CA348103967 rs1267981472 |
98 | E>* | No |
ClinGen TOPMed |
|
|
CA348103970 rs1306556692 |
98 | E>G | No |
ClinGen TOPMed |
|
|
CA1815086 rs763135850 |
99 | Y>S | No |
ClinGen ExAC gnomAD |
|
|
rs764192536 CA1815087 |
101 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764192536 CA348103987 |
101 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815088 rs774367973 |
102 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA348103999 rs761674486 |
103 | G>C | No |
ClinGen ExAC gnomAD |
|
|
CA1815089 rs761674486 |
103 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs767425900 CA1815090 |
104 | S>R | No |
ClinGen ExAC gnomAD |
|
|
COSM1527008 rs1335454177 CA348104013 COSM1527009 |
105 | G>S | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs755994999 CA1815092 |
106 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1573244235 CA348104025 |
107 | D>A | No |
ClinGen Ensembl |
|
|
CA348104042 rs1391915772 |
109 | I>M | No |
ClinGen gnomAD |
|
|
rs1398472942 CA348104039 |
109 | I>N | No |
ClinGen gnomAD |
|
|
CA348104045 rs1285252783 |
110 | Y>D | No |
ClinGen gnomAD |
|
|
rs766256305 CA348104052 |
111 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815093 rs766256305 |
111 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753586078 CA1815094 |
111 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs778588738 CA1815096 |
113 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA348104072 rs1417113083 |
114 | I>V | No |
ClinGen TOPMed |
|
|
CA1815099 rs781754645 |
115 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs569250884 CA53770661 |
116 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
CA348104086 rs569250884 |
116 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA348104090 rs1235034975 |
117 | F>Y | No |
ClinGen TOPMed |
|
|
rs199727155 CA53770667 |
120 | F>L | No |
ClinGen ExAC |
|
|
CA348104117 rs1359678741 |
121 | A>D | No |
ClinGen TOPMed |
|
|
CA1815103 rs749536605 |
121 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815104 rs749536605 |
121 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1200674888 CA348104125 |
122 | Y>F | No |
ClinGen gnomAD |
|
|
CA1815107 rs772032136 |
123 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs147879012 CA1815110 |
125 | E>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA348104141 rs1298737987 |
125 | E>G | No |
ClinGen TOPMed |
|
|
rs147879012 CA1815109 |
125 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs147879012 CA348104139 |
125 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs375422663 CA1815112 |
126 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1815113 rs200636557 |
127 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1394316248 CA348104150 |
127 | E>Q | No |
ClinGen gnomAD |
|
|
CA348104191 rs1426817880 |
133 | V>L | No |
ClinGen TOPMed |
|
|
rs937139915 CA53770710 |
136 | S>L | No |
ClinGen Ensembl |
|
|
CA1815116 rs777123504 |
137 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348104220 rs1297768575 |
138 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs756566928 CA1815118 |
140 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs892782902 CA348104240 |
141 | M>K | No |
ClinGen TOPMed |
|
|
CA53770742 rs892782902 |
141 | M>R | No |
ClinGen TOPMed |
|
|
rs1225120822 CA348104272 |
145 | S>N | No |
ClinGen TOPMed |
|
|
rs749591659 CA1815120 |
150 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1491003245 CA348104310 |
150 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA1815121 rs769007984 |
153 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA53770758 rs769007984 |
153 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
CA348104341 rs1196796618 |
155 | G>R | No |
ClinGen gnomAD |
|
|
CA1815123 rs748295860 |
157 | I>N | No |
ClinGen ExAC gnomAD |
|
|
CA348104361 rs1169212495 |
158 | G>S | No |
ClinGen gnomAD |
|
|
rs1371198261 CA348104373 |
159 | W>* | No |
ClinGen gnomAD |
|
|
rs931923616 CA53770766 |
160 | F>V | No |
ClinGen Ensembl |
|
|
rs1426494010 CA348104394 |
162 | S>F | No |
ClinGen gnomAD |
|
|
CA348104411 rs372651078 |
165 | V>F | No |
ClinGen ESP ExAC gnomAD |
|
|
CA1815127 rs372651078 |
165 | V>I | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1223200437 CA348104420 |
166 | L>W | No |
ClinGen TOPMed |
|
|
CA1815128 rs776572130 |
168 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA348104446 rs1284994943 |
170 | D>N | No |
ClinGen TOPMed |
|
|
CA348104458 rs1405256560 |
171 | E>D | No |
ClinGen TOPMed |
|
|
CA348104463 rs1362156483 |
172 | A>E | No |
ClinGen gnomAD |
|
|
CA1815130 rs765106638 |
174 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA1815132 rs762479770 |
175 | E>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs11553852 CA53770837 |
176 | S>P | No |
ClinGen Ensembl |
|
|
rs369379486 CA1815133 |
176 | S>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1178579083 CA348104490 |
177 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA348104493 rs1243481771 |
177 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA1815134 rs750977650 |
178 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA348104500 rs1419024844 |
178 | S>R | No |
ClinGen TOPMed |
|
|
CA1815135 rs756622014 |
179 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs534727383 CA1815136 |
180 | L>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA348104523 rs1573244918 |
182 | L>P | No |
ClinGen Ensembl |
|
|
rs1468183226 CA348104527 |
183 | R>C | No |
ClinGen gnomAD |
|
|
rs754330998 CA1815137 |
183 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754330998 CA348104528 |
183 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs373413217 CA53770858 |
185 | G>S | No |
ClinGen ESP |
|
|
CA53770859 rs755385208 |
186 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755385208 CA1815138 |
186 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779117347 CA1815139 |
187 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777919935 CA1815142 |
188 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1479928639 CA348104565 |
190 | N>D | No |
ClinGen gnomAD |
|
|
rs747018579 CA1815143 |
190 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA1815146 rs201888318 |
192 | Q>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs771183263 CA53770885 |
192 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
rs771183263 CA1815144 |
192 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs376669820 CA1815147 |
193 | G>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA348104592 rs1264859840 |
194 | S>F | No |
ClinGen TOPMed |
|
|
rs763589848 CA1815150 |
196 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs762683461 CA348104600 |
196 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762683461 COSM2981295 COSM2981294 CA1815149 |
196 | V>M | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA1815151 rs773992501 |
197 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA1815154 rs754384046 |
198 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA1815153 COSM1398598 COSM1398599 rs766993432 |
198 | H>Y | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC |
|
CA1815155 rs755440418 |
202 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs752945323 CA1815157 |
205 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA1815158 rs758649473 |
208 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA1815159 rs777924793 |
208 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA1815161 rs757457379 |
209 | V>F | No |
ClinGen ExAC gnomAD |
|
|
CA348104684 rs1445013870 |
210 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
CA348104692 rs1573245210 |
211 | E>D | No |
ClinGen Ensembl |
|
|
COSM1004788 CA53770991 COSM1004787 rs776751269 |
211 | E>K | endometrium [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA348104694 rs1373018176 |
212 | E>K | No |
ClinGen gnomAD |
|
|
rs745828163 CA1815163 |
213 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA1815165 rs775361933 |
215 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA348104770 rs761423872 |
222 | M>R | No |
ClinGen ExAC gnomAD |
|
|
CA1815169 rs761423872 |
222 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA53771015 rs972066107 |
223 | E>K | No |
ClinGen TOPMed |
|
|
CA53771045 rs1005968215 |
225 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1429091158 CA348104797 |
226 | E>G | No |
ClinGen TOPMed |
|
|
CA348104814 rs1179830468 |
228 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA348104812 rs1179830468 |
228 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1197655713 CA348104809 |
228 | P>T | No |
ClinGen TOPMed |
|
|
CA53771046 rs878946953 |
229 | E>K | No |
ClinGen Ensembl |
|
|
rs1175736952 CA348104867 |
235 | W>C | No |
ClinGen gnomAD |
|
|
rs1221973306 CA348104897 |
239 | N>T | No |
ClinGen gnomAD |
|
|
rs765594937 CA1815174 |
240 | A>P | No |
ClinGen ExAC TOPMed |
|
|
rs765594937 CA348104902 |
240 | A>S | No |
ClinGen ExAC TOPMed |
|
|
CA1815175 rs753243958 |
240 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA348104906 rs1411771322 |
241 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs758746326 CA1815176 |
241 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA348104907 rs1411771322 |
241 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs975509135 CA53771059 |
242 | G>S | No |
ClinGen Ensembl |
|
|
CA348104922 rs1440465020 |
243 | Q>H | No |
ClinGen gnomAD |
|
|
CA348104925 rs1322701308 |
244 | V>L | No |
ClinGen TOPMed |
|
|
rs1305346471 CA348104932 |
245 | G>D | No |
ClinGen gnomAD |
|
|
rs954576405 CA53771063 |
247 | V>I | No |
ClinGen gnomAD |
|
|
rs751867472 CA1815178 |
249 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348104980 rs1296634496 |
252 | V>G | No |
ClinGen TOPMed |
|
|
rs1573245524 CA348104983 |
253 | V>L | No |
ClinGen Ensembl |
|
|
rs757510743 CA1815179 |
254 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA53771081 rs371236589 |
255 | L>F | No |
ClinGen ESP gnomAD |
|
|
COSM144110 COSM1669192 COSM1669193 CA1815181 rs750464687 |
257 | D>E | large_intestine skin [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs779917800 CA1815183 |
258 | G>E | No |
ClinGen ExAC gnomAD |
|
|
rs756217972 CA1815182 |
258 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs137868721 CA1815185 |
259 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA348105027 rs778640285 |
261 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747822398 CA1815187 |
262 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA348105034 rs1165809099 |
262 | H>Y | No |
ClinGen gnomAD |
|
|
CA1815189 rs772711255 |
263 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA348105049 rs1479873527 |
264 | A>V | No |
ClinGen gnomAD |
|
|
CA53771127 rs990070802 |
265 | H>D | No |
ClinGen TOPMed |
|
|
CA1815192 rs377759138 |
265 | H>Q | No |
ClinGen ESP ExAC gnomAD |
|
|
rs770460763 CA1815191 |
265 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348105055 rs763482146 |
266 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815193 rs763482146 |
266 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815194 rs764581321 |
268 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA348105070 rs1226124084 |
268 | Q>R | No |
ClinGen gnomAD |
|
|
CA1815195 rs751920917 |
270 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA1815196 rs762133170 |
271 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs947129132 CA53771166 |
272 | T>S | No |
ClinGen TOPMed |
|
|
CA53771168 rs750648011 |
273 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815198 rs750648011 |
273 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1573245763 CA348105105 |
274 | P>S | No |
ClinGen Ensembl |
|
|
rs902885784 CA53771172 |
275 | S>W | No |
ClinGen TOPMed gnomAD |
|
|
CA53771177 rs549114543 |
277 | S>T | No |
ClinGen Ensembl |
|
|
rs1480875247 CA348105135 |
279 | R>C | No |
ClinGen TOPMed |
|
|
CA1815200 rs780151498 COSM1004791 COSM1004793 |
279 | R>H | large_intestine endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs374552067 CA1815201 |
280 | F>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA348105142 rs1386468934 |
280 | F>S | No |
ClinGen gnomAD |
|
|
CA53771191 rs938578667 |
281 | A>G | No |
ClinGen TOPMed |
|
|
rs1382720762 CA348105155 |
282 | G>V | No |
ClinGen TOPMed |
|
|
rs1388052573 CA348105158 |
283 | R>K | No |
ClinGen gnomAD |
|
|
rs1402102793 CA348105164 |
284 | E>Q | No |
ClinGen gnomAD |
|
|
rs1573245898 CA348105173 |
285 | W>G | No |
ClinGen Ensembl |
|
|
CA53771197 rs894285243 |
286 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
CA348105188 rs1397706082 |
287 | Y>H | No |
ClinGen TOPMed |
|
|
rs1383526320 CA348105207 |
289 | N>K | No |
ClinGen TOPMed gnomAD |
|
|
CA1815205 rs771849479 |
289 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs199651832 COSM1631232 CA53771202 COSM1631231 |
290 | V>M | liver [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs370241867 CA53771207 |
291 | T>M | No |
ClinGen ESP TOPMed |
|
|
CA348105223 rs1205980457 |
293 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA1815208 rs770384522 |
294 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA348105250 rs1183954324 |
296 | E>D | No |
ClinGen TOPMed |
|
|
rs769226018 CA348105259 |
297 | C>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1244215593 CA348105267 |
299 | L>F | No |
ClinGen gnomAD |
|
|
rs1444575856 CA348105279 |
300 | N>K | No |
ClinGen gnomAD |
|
|
CA1815212 rs774996531 |
301 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs767975664 CA348105298 |
304 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767975664 CA1815214 |
304 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA348105346 rs1294156140 |
311 | R>G | No |
ClinGen TOPMed |
|
|
rs765223593 CA1815220 |
312 | D>A | No |
ClinGen ExAC gnomAD |
|
|
rs368417738 CA1815219 |
312 | D>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs368417738 CA1815218 |
312 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs950214307 CA53771306 |
316 | S>A | No |
ClinGen Ensembl |
|
|
rs577283510 CA1815246 |
318 | S>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs748686837 CA348105412 |
318 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA348105411 rs748686837 |
318 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA1815249 rs773455664 |
319 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA1815248 rs772650775 |
319 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs904856726 CA53775347 |
320 | F>L | No |
ClinGen TOPMed |
|
|
rs1273249659 CA348105432 |
321 | S>C | No |
ClinGen TOPMed |
|
|
CA1815250 rs200594599 |
322 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA348105469 rs1490052739 |
326 | A>T | No |
ClinGen gnomAD |
|
|
CA348105474 COSM1004795 COSM1004794 rs1332586401 |
326 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1270571800 CA348105484 |
328 | G>E | No |
ClinGen gnomAD |
|
|
CA1815254 rs765331885 |
331 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA53775367 rs895059179 |
335 | V>M | No |
ClinGen TOPMed |
|
|
rs775496029 CA1815255 |
336 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA1815256 rs147922486 |
338 | V>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1815257 rs764022792 |
340 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs751441679 CA1815258 |
340 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs375398048 CA1815259 |
341 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1428631760 CA348105581 |
342 | Y>F | No |
ClinGen gnomAD |
|
|
rs749337403 CA53775378 |
344 | I>T | No |
ClinGen Ensembl |
|
|
COSM1398601 CA1815261 COSM1398602 rs750135733 |
348 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1476316800 CA348105619 |
348 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs750135733 CA348105617 |
348 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA1815263 rs779724766 |
350 | H>L | No |
ClinGen ExAC gnomAD |
|
|
rs77614409 CA1815264 |
351 | T>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1456606773 CA348105659 |
354 | E>K | No |
ClinGen gnomAD |
|
|
CA1815266 rs542899610 |
360 | K>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1815267 rs747449570 |
362 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771366302 CA1815268 |
362 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA348105754 rs1312487716 |
367 | S>R | No |
ClinGen gnomAD |
|
|
rs775551208 CA1815272 |
368 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs763085878 CA1815273 |
369 | H>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1815274 rs768598254 |
370 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA348105770 rs768598254 |
370 | G>W | No |
ClinGen ExAC gnomAD |
|
|
CA1815278 rs144762403 |
374 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA53775416 rs1026995769 |
374 | Y>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1412845098 CA348105807 |
375 | L>P | No |
ClinGen gnomAD |
|
|
CA348105809 rs1161526080 |
376 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA348105817 rs1456773337 |
377 | R>K | No |
ClinGen gnomAD |
|
|
rs1377398900 CA348105833 |
380 | Q>K | No |
ClinGen gnomAD |
No associated diseases with O43639
8 regional properties for O43639
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH2 domain | 283 - 379 | IPR000980 |
| domain | SH3 domain | 2 - 61 | IPR001452-1 |
| domain | SH3 domain | 111 - 170 | IPR001452-2 |
| domain | SH3 domain | 195 - 257 | IPR001452-3 |
| domain | Nck2, SH3 domain 1 | 2 - 59 | IPR035559 |
| domain | Nck2, SH3 domain 2 | 114 - 168 | IPR035560 |
| domain | Nck2, SH3 domain 3 | 198 - 256 | IPR035561 |
| domain | Nck2, SH2 domain | 283 - 380 | IPR035883 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR19969 | SH2-SH3 ADAPTOR PROTEIN-RELATED |
| PANTHER Subfamily | PTHR19969:SF12 | CYTOPLASMIC PROTEIN NCK2 |
| PANTHER Protein Class | scaffold/adaptor protein | |
| PANTHER Pathway Category |
Angiogenesis Nck PDGF signaling pathway Nck T cell activation nck |
|
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| vesicle membrane | The lipid bilayer surrounding any membrane-bounded vesicle in the cell. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| cytoskeletal anchor activity | The binding activity of a protein that brings together a cytoskeletal protein (either a microtubule or actin filament, spindle pole body, or protein directly bound to them) and one or more other molecules, permitting them to function in a coordinated way. |
| phosphotyrosine residue binding | Binding to a phosphorylated tyrosine residue within a protein. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| scaffold protein binding | Binding to a scaffold protein. Scaffold proteins are crucial regulators of many key signaling pathways. Although not strictly defined in function, they are known to interact and/or bind with multiple members of a signaling pathway, tethering them into complexes. |
| signaling adaptor activity | The binding activity of a molecule that brings together two or more molecules in a signaling pathway, permitting those molecules to function in a coordinated way. Adaptor molecules themselves do not have catalytic activity. |
| signaling receptor complex adaptor activity | The binding activity of a molecule that provides a physical support for the assembly of a multiprotein receptor signaling complex. |
21 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| dendritic spine development | The process whose specific outcome is the progression of the dendritic spine over time, from its formation to the mature structure. A dendritic spine is a protrusion from a dendrite and a specialized subcellular compartment involved in synaptic transmission. |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| epidermal growth factor receptor signaling pathway | The series of molecular signals initiated by binding of a ligand to the tyrosine kinase receptor EGFR (ERBB1) on the surface of a cell. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| immunological synapse formation | The formation of an area of close contact between a lymphocyte (T-, B-, or natural killer cell) and a target cell through the clustering of particular signaling and adhesion molecules and their associated membrane rafts on both the lymphocyte and target cell, which facilitates activation of the lymphocyte, transfer of membrane from the target cell to the lymphocyte, and in some situations killing of the target cell through release of secretory granules and/or death-pathway ligand-receptor interaction. |
| lamellipodium assembly | Formation of a lamellipodium, a thin sheetlike extension of the surface of a migrating cell. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | Any process that stops, prevents or reduces the frequency, rate or extent of endoplasmic reticulum stress-induced eiF2alpha phosphorylation. |
| negative regulation of peptidyl-serine phosphorylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the phosphorylation of peptidyl-serine. |
| negative regulation of PERK-mediated unfolded protein response | Any process that stops, prevents or reduces the frequency, rate or extent of the PERK-mediated unfolded protein response. |
| negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription from an RNA polymerase II promoter as a result of an endoplasmic reticulum stress. |
| positive regulation of actin filament polymerization | Any process that activates or increases the frequency, rate or extent of actin polymerization. |
| positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | Any process that activates or increases the frequency, rate or extent of an endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway. |
| positive regulation of T cell proliferation | Any process that activates or increases the rate or extent of T cell proliferation. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of translation in response to endoplasmic reticulum stress | Any process that activates, or increases the frequency, rate or extent of translation as a result of endoplasmic reticulum stress. |
| regulation of epidermal growth factor-activated receptor activity | Any process that modulates the frequency, rate or extent of EGF-activated receptor activity. |
| signal complex assembly | The aggregation, arrangement and bonding together of a set of components to form a complex capable of relaying a signal within a cell. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| T cell activation | The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P16333 | NCK1 | Cytoplasmic protein NCK1 | Homo sapiens (Human) | PR |
| P46108 | CRK | Adapter molecule crk | Homo sapiens (Human) | EV |
| P46109 | CRKL | Crk-like protein | Homo sapiens (Human) | SS |
| Q99M51 | Nck1 | Cytoplasmic protein NCK1 | Mus musculus (Mouse) | PR |
| O55033 | Nck2 | Cytoplasmic protein NCK2 | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTEEVIVIAK | WDYTAQQDQE | LDIKKNERLW | LLDDSKTWWR | VRNAANRTGY | VPSNYVERKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SLKKGSLVKN | LKDTLGLGKT | RRKTSARDAS | PTPSTDAEYP | ANGSGADRIY | DLNIPAFVKF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AYVAEREDEL | SLVKGSRVTV | MEKCSDGWWR | GSYNGQIGWF | PSNYVLEEVD | EAAAESPSFL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SLRKGASLSN | GQGSRVLHVV | QTLYPFSSVT | EEELNFEKGE | TMEVIEKPEN | DPEWWKCKNA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RGQVGLVPKN | YVVVLSDGPA | LHPAHAPQIS | YTGPSSSGRF | AGREWYYGNV | TRHQAECALN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ERGVEGDFLI | RDSESSPSDF | SVSLKASGKN | KHFKVQLVDN | VYCIGQRRFH | TMDELVEHYK |
| 370 | |||||
| KAPIFTSEHG | EKLYLVRALQ |