O43633
Gene name |
CHMP2A (BC2, CHMP2) |
Protein name |
Charged multivesicular body protein 2a |
Names |
Chromatin-modifying protein 2a, CHMP2a, Putative breast adenocarcinoma marker BC-2, Vacuolar protein sorting-associated protein 2-1, Vps2-1, hVps2-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:27243 |
EC number |
|
Protein Class |
CHARGED MULTIVESICULAR BODY PROTEIN (PTHR10476) |
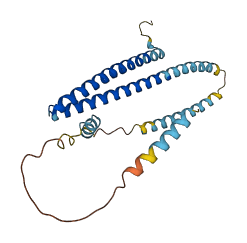
Descriptions
CHMP2A is a core component of the endosomal sorting required for transport complex III (ESCRT-III), which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. Removing 42 amino acids from the C-terminal of the protein unmasks a common ability to associate with endosomal membranes and assemble into large polymeric complexes, and the C-terminal sequences are also important for interaction of ESCRT-III protein with VPS4. Thus, ESCRT-III proteins cycle between a closed state and an activated open state is under control of sequences at the C-terminus and associated factors.
Autoinhibitory domains (AIDs)
Target domain |
1-144 (N-terminal α1-α4 domains) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

3 structures for O43633
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 7ZCG | EM | 330 A | B | 8-154 | PDB |
| 7ZCH | EM | 360 A | B | 8-154 | PDB |
| AF-O43633-F1 | Predicted | AlphaFoldDB |
185 variants for O43633
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1255752892 CA407935987 |
3 | L>Q | No |
ClinGen TOPMed |
|
|
CA310614461 rs981971580 |
4 | L>W | No |
ClinGen Ensembl |
|
|
CA9719877 rs758966430 |
5 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA407935899 rs1166533738 |
6 | G>R | No |
ClinGen gnomAD |
|
|
CA407935880 rs1568668219 |
7 | R>C | No |
ClinGen Ensembl |
|
|
rs949340234 CA310614459 |
8 | R>P | No |
ClinGen TOPMed |
|
|
CA310614456 rs1044565 |
13 | E>G | No |
ClinGen Ensembl |
|
|
CA407935656 rs1239326026 |
13 | E>K | No |
ClinGen gnomAD |
|
|
rs773101288 CA9719873 |
15 | L>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1568668148 CA407935548 |
17 | Q>L | No |
ClinGen Ensembl |
|
|
rs1600089259 CA407935512 |
18 | N>S | No |
ClinGen Ensembl |
|
|
CA407935476 rs1222634863 |
19 | Q>R | No |
ClinGen gnomAD |
|
|
CA407935418 rs1284040289 |
21 | A>T | No |
ClinGen gnomAD |
|
|
CA310614449 rs1023876786 |
26 | M>T | No |
ClinGen Ensembl |
|
|
rs866674822 CA310614448 |
27 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs775433926 CA9719870 |
28 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA407935115 rs1218523927 |
30 | D>N | No |
ClinGen gnomAD |
|
|
CA407935101 rs1365279794 |
30 | D>V | No |
ClinGen gnomAD |
|
|
rs748088651 CA310614445 |
31 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA9719869 rs748088651 |
31 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs1436829673 CA407935074 |
31 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA9719868 rs745916847 |
34 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA310614442 rs893544316 |
37 | E>Q | No |
ClinGen TOPMed |
|
|
rs757434481 CA9719865 |
38 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA407934793 rs1176041886 |
39 | Q>* | No |
ClinGen gnomAD |
|
|
rs1338551455 CA407934758 |
40 | E>K | No |
ClinGen TOPMed |
|
|
CA310614438 rs776623849 |
40 | E>V | No |
ClinGen Ensembl |
|
|
CA407934564 rs1192124915 |
42 | K>T | No |
ClinGen gnomAD |
|
|
CA310614434 rs1055130374 |
44 | I>T | No |
ClinGen TOPMed |
|
|
CA9719862 rs778055563 |
44 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA9719859 rs779276855 |
47 | I>N | No |
ClinGen ExAC |
|
|
rs753315789 CA9719860 |
47 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs548744646 CA9719857 |
52 | K>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA310614430 rs937989967 |
52 | K>R | No |
ClinGen TOPMed |
|
|
CA9719855 rs766161099 |
54 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1339557310 CA407933939 |
56 | M>I | No |
ClinGen gnomAD |
|
|
CA407933981 rs1223901907 |
56 | M>V | No |
ClinGen gnomAD |
|
|
CA407932255 rs1457256423 |
57 | D>H | No |
ClinGen gnomAD |
|
|
CA9719827 rs760731461 |
59 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs773474943 CA310613542 |
60 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773474943 CA9719826 |
60 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772152550 CA9719825 COSM1743009 |
60 | R>H | biliary_tract [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs773474943 CA310613544 |
60 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1045368903 CA310613539 |
62 | M>I | No |
ClinGen Ensembl |
|
|
rs1242100512 CA407932093 |
64 | K>E | No |
ClinGen gnomAD |
|
|
rs779096961 CA9719822 |
66 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs748574916 CA9719823 |
66 | L>S | No |
ClinGen ExAC gnomAD |
|
|
rs1216396709 CA407931996 |
67 | V>A | No |
ClinGen TOPMed |
|
|
CA9719821 COSM1681475 rs769219232 |
68 | R>C | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ExAC |
|
COSM714031 rs749741448 CA9719820 |
68 | R>H | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA9719819 rs369929153 |
69 | T>N | No |
ClinGen ESP ExAC gnomAD |
|
|
rs755740005 CA9719818 |
70 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1466949169 CA407931959 |
70 | R>W | No |
ClinGen TOPMed |
|
|
rs781025283 CA9719816 |
71 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs376979258 CA9719814 COSM274538 |
74 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA9719813 COSM1641264 rs763733145 |
74 | R>H | stomach [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs763733145 CA407931838 |
74 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA9719811 rs758208886 |
75 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA310613529 rs950335550 |
77 | V>E | No |
ClinGen Ensembl |
|
|
rs916170048 CA310613528 |
78 | L>S | No |
ClinGen Ensembl |
|
|
CA407931705 rs146786448 |
80 | R>G | No |
ClinGen ESP gnomAD |
|
|
rs765137954 CA9719809 |
80 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs146786448 CA310613526 |
80 | R>W | No |
ClinGen ESP gnomAD |
|
|
CA407931567 rs1419062527 |
84 | Q>R | No |
ClinGen TOPMed |
|
|
CA310613524 rs529552077 |
86 | V>M | No |
ClinGen Ensembl |
|
|
rs773233623 CA9719807 |
87 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA310613522 rs753745398 |
87 | S>P | No |
ClinGen Ensembl |
|
|
rs774782757 CA9719805 |
89 | K>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9719804 rs774782757 |
89 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9719802 rs143635425 |
91 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1325589161 CA407931291 |
92 | T>A | No |
ClinGen TOPMed |
|
|
CA407931239 rs1568666405 |
93 | L>I | No |
ClinGen Ensembl |
|
|
rs1321987514 CA407931215 |
94 | K>E | No |
ClinGen gnomAD |
|
|
rs1388799491 CA407931163 |
95 | S>F | No |
ClinGen gnomAD |
|
|
CA407931084 rs1297745371 |
97 | N>S | No |
ClinGen gnomAD |
|
|
rs1438168135 CA407931063 |
98 | S>A | No |
ClinGen TOPMed |
|
|
CA9719801 rs776185223 |
98 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA407931026 rs745397145 |
99 | M>R | No |
ClinGen ExAC gnomAD |
|
|
CA9719799 rs745397145 |
99 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA407931002 rs1470453514 |
100 | A>T | No |
ClinGen gnomAD |
|
|
rs780514546 CA9719798 |
101 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA9719797 rs756942841 |
105 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1434621947 CA407930770 |
105 | G>V | No |
ClinGen gnomAD |
|
|
rs746707272 CA9719796 |
111 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs1228975817 CA407930512 |
111 | G>S | No |
ClinGen TOPMed |
|
|
CA310613512 rs957576851 |
112 | T>I | No |
ClinGen Ensembl |
|
|
CA407930378 rs1206311342 |
113 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1206311342 CA407930375 |
113 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
CA407930386 rs1287504375 |
113 | M>T | No |
ClinGen gnomAD |
|
|
rs1262462964 CA407930353 |
114 | N>D | No |
ClinGen gnomAD |
|
|
rs1235222910 CA407929967 |
121 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1271180892 CA407929944 |
122 | I>M | No |
ClinGen TOPMed |
|
|
CA9719772 rs754914078 |
124 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA407929764 rs1320400002 |
126 | M>I | No |
ClinGen gnomAD |
|
|
CA407929790 rs1197159892 |
126 | M>R | No |
ClinGen TOPMed gnomAD |
|
|
CA407929750 rs1274002003 |
127 | M>T | No |
ClinGen gnomAD |
|
|
CA407929687 rs1254068409 |
128 | E>D | No |
ClinGen TOPMed |
|
|
CA9719770 rs753785301 COSM1236597 |
131 | R>Q | autonomic_ganglia [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs960083867 CA310613486 |
131 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs1383450597 CA407929497 |
134 | E>K | No |
ClinGen gnomAD |
|
|
rs1238456522 CA407929438 |
135 | I>V | No |
ClinGen TOPMed |
|
|
rs757326974 CA9719768 |
136 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA310613482 rs980225414 |
136 | M>K | No |
ClinGen TOPMed gnomAD |
|
|
CA407929375 rs980225414 |
136 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
CA9719767 rs751625630 |
137 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA407929321 rs751625630 |
137 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA310613479 rs970335404 |
138 | M>I | No |
ClinGen TOPMed |
|
|
CA407929266 rs1404363876 |
138 | M>T | No |
ClinGen gnomAD |
|
|
CA407929270 rs1418369334 |
138 | M>V | No |
ClinGen TOPMed |
|
|
rs1411082318 CA407929227 |
139 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1027639495 CA310613478 |
139 | K>T | No |
ClinGen TOPMed |
|
|
rs1600086997 CA407929201 |
140 | E>K | No |
ClinGen Ensembl |
|
|
rs1380258678 CA407929101 |
141 | E>A | No |
ClinGen gnomAD |
|
|
CA9719763 rs753120384 |
142 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA9719761 rs759965981 |
143 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1422972057 CA407928960 |
143 | M>K | No |
ClinGen gnomAD |
|
|
rs765768118 CA9719762 |
143 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9719760 rs777054240 |
145 | D>Y | No |
ClinGen ExAC |
|
|
CA407928801 rs1246804598 |
146 | A>T | No |
ClinGen gnomAD |
|
|
CA9719758 rs760136138 |
147 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA310613470 rs369433950 |
149 | D>A | No |
ClinGen ESP gnomAD |
|
|
rs1341808043 CA407928508 |
152 | G>D | No |
ClinGen TOPMed |
|
|
CA407928516 rs1435388230 |
152 | G>S | No |
ClinGen gnomAD |
|
|
rs1028258425 CA310613467 |
153 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1600086915 CA407928347 |
156 | D>E | No |
ClinGen Ensembl |
|
|
rs749084452 CA9719752 |
156 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs749084452 CA9719751 |
156 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs375168365 CA9719749 |
158 | E>D | No |
ClinGen ESP ExAC TOPMed |
|
|
COSM3797649 rs1440468684 CA407928294 |
158 | E>K | urinary_tract [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs751607738 CA407928248 |
159 | E>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9719748 rs751607738 |
159 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA407928228 rs1409245486 |
159 | E>V | No |
ClinGen gnomAD |
|
|
rs1241885185 CA407928188 |
160 | S>T | No |
ClinGen TOPMed |
|
|
CA9719719 rs755182193 |
162 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs146139219 CA9719718 |
164 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs187800344 CA310613432 |
165 | S>A | No |
ClinGen 1000Genomes gnomAD |
|
|
CA407927837 rs1237576842 |
165 | S>F | No |
ClinGen TOPMed |
|
|
CA310613431 rs936234835 |
166 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA310613428 rs201891984 |
170 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1255518400 CA407927578 |
172 | G>V | No |
ClinGen gnomAD |
|
|
CA407927542 rs1181275546 |
173 | L>R | No |
ClinGen gnomAD |
|
|
rs773810226 CA9719712 |
174 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9719713 rs773810226 |
174 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9719710 rs762601677 |
175 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs775002155 CA9719709 |
177 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs745628642 CA9719708 |
178 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA407927391 rs1229134555 |
178 | E>K | No |
ClinGen gnomAD |
|
|
CA310613420 rs977579289 |
179 | L>V | No |
ClinGen Ensembl |
|
|
CA9719706 rs144804005 COSM1002788 |
180 | S>L | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs778121638 CA310613388 |
182 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs778121638 CA9719675 |
182 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs891673180 CA310613385 |
185 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1396990938 CA407926956 |
185 | T>P | No |
ClinGen TOPMed |
|
|
CA9719673 rs752113260 |
187 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs149478183 CA9719674 |
187 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs541443283 CA9719671 |
188 | S>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs760530137 CA9719668 |
189 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA9719667 rs773247293 |
190 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs1255363145 CA407926616 |
192 | A>T | No |
ClinGen TOPMed |
|
|
CA407926581 rs1454141226 |
193 | A>G | No |
ClinGen gnomAD |
|
|
rs556423277 CA9719666 |
193 | A>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA310613379 rs938655319 |
194 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs938655319 CA407926576 |
194 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA9719665 rs183266343 |
195 | G>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA310613377 rs76948000 |
196 | K>E | No |
ClinGen Ensembl |
|
|
rs1431733496 CA407926425 |
198 | A>T | No |
ClinGen TOPMed |
|
|
CA9719663 rs770015701 |
199 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1467001606 CA407926319 |
200 | A>S | No |
ClinGen TOPMed |
|
|
CA9719660 rs138058612 |
201 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM290638 CA407926231 rs1568665125 |
202 | A>S | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs974233538 CA310613373 |
203 | S>L | No |
ClinGen TOPMed |
|
|
rs200224355 CA9719659 |
204 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA407926120 rs1277732919 |
204 | A>V | No |
ClinGen gnomAD |
|
|
rs1600086200 CA407926080 |
205 | L>P | No |
ClinGen Ensembl |
|
|
CA9719658 rs553668807 |
206 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs368139604 CA9719657 |
207 | D>H | No |
ClinGen ESP ExAC gnomAD |
|
|
CA407925927 rs1382682787 |
208 | A>D | No |
ClinGen gnomAD |
|
|
CA407925872 rs1405154683 |
209 | D>V | No |
ClinGen TOPMed gnomAD |
|
|
rs778221913 CA9719655 |
210 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA407925729 rs1429593090 |
214 | E>K | No |
ClinGen gnomAD |
|
|
CA407925579 rs1600086104 |
218 | N>T | No |
ClinGen Ensembl |
|
|
rs756008421 CA9719651 |
220 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9719652 rs766103995 |
220 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs904782704 CA310613366 |
222 | D>N | No |
ClinGen Ensembl |
|
|
CA9719649 rs767511825 |
223 | D>W | No |
ClinGen ExAC |
No associated diseases with O43633
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR10476 | CHARGED MULTIVESICULAR BODY PROTEIN |
| PANTHER Subfamily | PTHR10476:SF4 | CHARGED MULTIVESICULAR BODY PROTEIN 2A |
| PANTHER Protein Class | membrane traffic protein | |
| PANTHER Pathway Category | No pathway information available | |
16 GO annotations of cellular component
| Name | Definition |
|---|---|
| amphisome membrane | Any membrane that is part of an amphisome. |
| autophagosome membrane | The lipid bilayer surrounding an autophagosome, a double-membrane-bounded vesicle in which endogenous cellular material is sequestered. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| ESCRT III complex | A complex with membrane scission activity that plays a major role in many processes where membranes are remodelled - including endosomal transport (vesicle budding), nuclear envelope organisation (membrane closure, mitotic bridge cleavage), and cytokinesis (abscission). |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| kinetochore | A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules. |
| kinetochore microtubule | Any of the spindle microtubules that attach to the kinetochores of chromosomes by their plus ends, and maneuver the chromosomes during mitotic or meiotic chromosome segregation. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| membrane coat | Any of several different proteinaceous coats that can associate with membranes. Membrane coats include those formed by clathrin plus an adaptor complex, the COPI and COPII complexes, and possibly others. They are found associated with membranes on many vesicles as well as other membrane features such as pits and perhaps tubules. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| multivesicular body | A type of endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body membrane | The lipid bilayer surrounding a multivesicular body. |
| nuclear envelope | The double lipid bilayer enclosing the nucleus and separating its contents from the rest of the cytoplasm; includes the intermembrane space, a gap of width 20-40 nm (also called the perinuclear space). |
| nuclear pore | A protein complex providing a discrete opening in the nuclear envelope of a eukaryotic cell, where the inner and outer nuclear membranes are joined. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| phosphatidylcholine binding | Binding to a phosphatidylcholine, a glycophospholipid in which a phosphatidyl group is esterified to the hydroxyl group of choline. |
| protein domain specific binding | Binding to a specific domain of a protein. |
29 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome maturation | Removal of PI3P and Atg8/LC3 after the closure of the phagophore and before the fusion with the endosome/lysosome (e.g. mammals and insects) or vacuole (yeast), and that very likely destabilizes other Atg proteins and thus enables their efficient dissociation and recycling. |
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| endosome transport via multivesicular body sorting pathway | The directed movement of substances from endosomes to lysosomes or vacuoles by a pathway in which molecules are sorted into multivesicular bodies, which then fuse with the target compartment. |
| ESCRT III complex disassembly | The disaggregation of an ESCRT III complex into its constituent components. |
| establishment of protein localization | The directed movement of a protein to a specific location. |
| exit from mitosis | The cell cycle transition where a cell leaves M phase and enters a new G1 phase. M phase is the part of the mitotic cell cycle during which mitosis and cytokinesis take place. |
| late endosome to lysosome transport | The directed movement of substances from late endosome to lysosome. |
| late endosome to vacuole transport | The directed movement of substances from late endosomes to the vacuole. In yeast, after transport to the prevacuolar compartment, endocytic content is delivered to the late endosome and on to the vacuole. This pathway is analogous to endosome to lysosome transport. |
| macroautophagy | The major inducible pathway for the general turnover of cytoplasmic constituents in eukaryotic cells, it is also responsible for the degradation of active cytoplasmic enzymes and organelles during nutrient starvation. Macroautophagy involves the formation of double-membrane-bounded autophagosomes which enclose the cytoplasmic constituent targeted for degradation in a membrane-bounded structure. Autophagosomes then fuse with a lysosome (or vacuole) releasing single-membrane-bounded autophagic bodies that are then degraded within the lysosome (or vacuole). Some types of macroautophagy, e.g. pexophagy, mitophagy, involve selective targeting of the targets to be degraded. |
| membrane invagination | The infolding of a membrane. |
| midbody abscission | The process by which the midbody, the cytoplasmic bridge that connects the two prospective daughter cells, is severed at the end of mitotic cytokinesis, resulting in two separate daughter cells. |
| mitotic metaphase plate congression | The cell cycle process in which chromosomes are aligned at the metaphase plate, a plane halfway between the poles of the mitotic spindle, during mitosis. |
| multivesicular body assembly | The aggregation, arrangement and bonding together of a set of components to form a multivesicular body, a type of late endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body sorting pathway | A vesicle-mediated transport process in which transmembrane proteins are ubiquitylated to facilitate their entry into luminal vesicles of multivesicular bodies (MVBs); upon subsequent fusion of MVBs with lysosomes or vacuoles, the cargo proteins are degraded. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of centriole elongation | Any process that stops, prevents or reduces the frequency, rate or extent of centriole elongation. |
| nuclear membrane reassembly | The reformation of the nuclear membranes following their breakdown in the context of a normal process. |
| nucleus organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the nucleus. |
| plasma membrane repair | The resealing of a cell plasma membrane after cellular wounding due to, for instance, mechanical stress. |
| positive regulation of exosomal secretion | Any process that activates or increases the frequency, rate or extent of exosomal secretion. |
| protein homooligomerization | The process of creating protein oligomers, compounds composed of a small number, usually between three and ten, of identical component monomers. Oligomers may be formed by the polymerization of a number of monomers or the depolymerization of a large protein polymer. |
| protein polymerization | The process of creating protein polymers, compounds composed of a large number of component monomers; polymeric proteins may be made up of different or identical monomers. Polymerization occurs by the addition of extra monomers to an existing poly- or oligomeric protein. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of centrosome duplication | Any process that modulates the frequency, rate or extent of centrosome duplication. Centrosome duplication is the replication of a centrosome, a structure comprised of a pair of centrioles and peri-centriolar material from which a microtubule spindle apparatus is organized. |
| regulation of mitotic spindle assembly | Any process that modulates the frequency, rate or extent of mitotic spindle assembly. |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | The chemical reactions and pathways resulting in the breakdown of a protein or peptide covalently tagged with ubiquitin, via the multivesicular body (MVB) sorting pathway; ubiquitin-tagged proteins are sorted into MVBs, and delivered to a lysosome/vacuole for degradation. |
| viral budding from plasma membrane | A viral budding that starts with formation of a membrane curvature in the host plasma membrane. |
| viral budding via host ESCRT complex | Viral budding which uses a host ESCRT protein complex, or complexes, to mediate the budding process. |
| viral release from host cell | The dissemination of mature viral particles from the host cell, e.g. by cell lysis or the budding of virus particles from the cell membrane. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5ZHN1 | CHMP2A | Charged multivesicular body protein 2a | Gallus gallus (Chicken) | SS |
| Q7LBR1 | CHMP1B | Charged multivesicular body protein 1b | Homo sapiens (Human) | PR |
| Q9Y3E7 | CHMP3 | Charged multivesicular body protein 3 | Homo sapiens (Human) | EV |
| Q9DB34 | Chmp2a | Charged multivesicular body protein 2a | Mus musculus (Mouse) | SS |
| Q9SKI2 | VPS2.1 | Vacuolar protein sorting-associated protein 2 homolog 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q6DFS6 | chmp2a | Charged multivesicular body protein 2a | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q7ZW25 | chmp2a | Charged multivesicular body protein 2a | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDLLFGRRKT | PEELLRQNQR | ALNRAMRELD | RERQKLETQE | KKIIADIKKM | AKQGQMDAVR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IMAKDLVRTR | RYVRKFVLMR | ANIQAVSLKI | QTLKSNNSMA | QAMKGVTKAM | GTMNRQLKLP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QIQKIMMEFE | RQAEIMDMKE | EMMNDAIDDA | MGDEEDEEES | DAVVSQVLDE | LGLSLTDELS |
| 190 | 200 | 210 | 220 | ||
| NLPSTGGSLS | VAAGGKKAEA | AASALADADA | DLEERLKNLR | RD |