O43320
Gene name |
FGF16 |
Protein name |
Fibroblast growth factor 16 |
Names |
FGF-16 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:8823 |
EC number |
|
Protein Class |
FIBROBLAST GROWTH FACTOR (PTHR11486) |
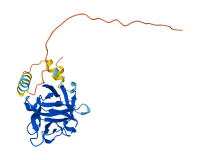
Descriptions
The mammalian fibroblast growth factor (FGF)1 family contains at least 22 distinct polypeptides (FGF1–FGF22) that are expressed in a specific spatial and temporal pattern. FGF9 was originally described as a glia-activating factor and is expressed in the nervous system as a potent mitogen for glia cells. FGF9 adopts a beta-trefoil fold similar to other FGFs, but the N- and C-terminal regions outside the beta-trefoil core are ordered (typical FGFs have disordered tails) and involved in reversible dimerization. Due to the dimerization, a significant surface area is buried in the dimer interface that occludes a major receptor binding site of FGF9 within the beta-trefoil core. Sequence alignment of FGFs shows that most of the residues that participate in this interface in FGF9 are conserved in FGF16, suggesting that FGF16 may also dimerize for autoinhibition.
Autoinhibitory domains (AIDs)
Target domain |
62-189 (beta-trefoil core) |
Relief mechanism |
|
Assay |
|
Target domain |
62-189 (beta-trefoil core) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for O43320
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O43320-F1 | Predicted | AlphaFoldDB |
131 variants for O43320
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
VAR_072396 rs2062545863 |
68 | R>L | MF4 [UniProt] | Yes |
TOPMed UniProt |
|
rs587777051 RCV000056297 CA144858 |
157 | S>* | Syndactyly type 8 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA144856 rs587777050 RCV000056296 |
179 | R>* | Variant assessed as Somatic; HIGH impact. Syndactyly type 8 [NCI-TCGA, ClinVar] | Yes |
ClinGen ClinVar NCI-TCGA TOPMed dbSNP gnomAD |
| rs2062545304 | 3 | E>K | No | gnomAD | |
| rs1339474563 | 4 | V>M | No |
TOPMed gnomAD |
|
| rs2062545372 | 5 | G>E | No | gnomAD | |
| rs1366904951 | 5 | G>R | No |
TOPMed gnomAD |
|
| rs2062545392 | 6 | G>D | No |
TOPMed gnomAD |
|
| rs2062545392 | 6 | G>V | No |
TOPMed gnomAD |
|
| rs934230464 | 9 | A>T | No | Ensembl | |
| rs2062545453 | 13 | W>* | No | Ensembl | |
| rs2062545445 | 13 | W>S | No | TOPMed | |
| rs2062545459 | 14 | D>V | No | TOPMed | |
| rs2147424275 | 15 | L>P | No | Ensembl | |
| rs1307694730 | 20 | S>L | No |
TOPMed gnomAD |
|
| rs1377332228 | 21 | S>F | No |
TOPMed gnomAD |
|
| rs2147424283 | 21 | S>T | No | Ensembl | |
| rs1433114171 | 23 | G>R | No | TOPMed | |
| rs1423758172 | 25 | V>A | No |
TOPMed gnomAD |
|
| rs2062545549 | 25 | V>L | No | Ensembl | |
| rs2062545581 | 28 | A>T | No | TOPMed | |
| rs1179264472 | 29 | D>V | No |
TOPMed gnomAD |
|
| rs1473127579 | 31 | P>S | No | TOPMed | |
| rs1203968737 | 35 | N>K | No |
TOPMed gnomAD |
|
| rs1481890060 | 36 | E>Q | No | TOPMed | |
| rs2147424325 | 39 | G>D | No | Ensembl | |
| rs1257236274 | 39 | G>S | No |
TOPMed gnomAD |
|
| rs1311475108 | 47 | R>C | No |
TOPMed gnomAD |
|
| rs1276419951 | 48 | G>C | No |
TOPMed gnomAD |
|
| rs2062545719 | 48 | G>D | No | gnomAD | |
| rs2062545754 | 49 | S>L | No |
1000Genomes TOPMed |
|
| rs1350909319 | 49 | S>T | No |
TOPMed gnomAD |
|
| rs2062545775 | 54 | A>V | No | gnomAD | |
| rs1303491352 | 61 | R>Q | No | TOPMed | |
| rs891850236 | 61 | R>W | No | Ensembl | |
| rs2062545839 | 62 | R>L | No |
TOPMed gnomAD |
|
| rs1357868650 | 73 | L>P | No |
TOPMed gnomAD |
|
| rs1161955059 | 75 | I>V | No | TOPMed | |
| rs1411645067 | 79 | G>C | No |
TOPMed gnomAD |
|
| rs1411645067 | 79 | G>S | No |
TOPMed gnomAD |
|
| rs2062545908 | 80 | T>A | No | Ensembl | |
| rs1180551153 | 80 | T>M | No |
TOPMed gnomAD |
|
| rs1471395875 | 83 | G>E | No |
TOPMed gnomAD |
|
| rs2062545927 | 83 | G>R | No | Ensembl | |
| rs2147424388 | 84 | T>I | No | Ensembl | |
| rs1257850910 | 85 | R>P | No | TOPMed | |
| rs1181865833 | 86 | H>R | No |
TOPMed gnomAD |
|
| rs1482660052 | 89 | S>N | No | TOPMed | |
| rs1235619458 | 90 | R>H | No | TOPMed | |
| rs2062565217 | 92 | G>E | No | TOPMed | |
| rs2062565299 | 96 | F>L | No | TOPMed | |
| rs1602277807 | 98 | S>N | No | Ensembl | |
| rs1401549641 | 104 | I>N | No | gnomAD | |
| rs781986970 | 106 | I>L | No |
ExAC gnomAD |
|
| rs782135458 | 107 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM3563489 rs188208804 |
107 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes NCI-TCGA TOPMed gnomAD |
|
COSM3563490 rs868993594 |
108 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs1420752953 | 108 | G>R | No | TOPMed | |
| COSM3845391 | 110 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 110 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1569510623 | 112 | G>V | No | Ensembl | |
| rs2147427123 | 119 | E>D | No | Ensembl | |
| rs1602277850 | 119 | E>G | No | Ensembl | |
| rs1192443222 | 120 | R>* | No | Ensembl | |
| rs1478286328 | 120 | R>L | No |
TOPMed gnomAD |
|
| rs1478286328 | 120 | R>Q | No |
TOPMed gnomAD |
|
| rs896431552 | 121 | G>A | No | Ensembl | |
| TCGA novel | 122 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 123 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1569510626 | 125 | G>A | No | Ensembl | |
| rs781972271 | 126 | S>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376876932 | 126 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP NCI-TCGA gnomAD |
| rs376876932 | 126 | S>W | No |
1000Genomes ESP gnomAD |
|
| rs2062571684 | 127 | K>E | No | Ensembl | |
| COSM1469425 | 127 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2062571708 | 129 | L>F | No | gnomAD | |
| rs1557027100 | 130 | T>I | No | gnomAD | |
| rs1247722394 | 131 | R>C | No |
TOPMed gnomAD |
|
| rs1557027102 | 131 | R>H | No |
TOPMed gnomAD |
|
| rs1557027102 | 131 | R>L | No |
TOPMed gnomAD |
|
|
rs782233147 COSM3783625 |
132 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA |
| rs2062571813 | 133 | C>Y | No | gnomAD | |
| rs1557027104 | 135 | F>S | No | gnomAD | |
|
rs782317053 COSM5757568 |
136 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| COSM356337 | 136 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781941470 | 137 | E>D | No |
ExAC gnomAD |
|
| rs2062571942 | 140 | E>Q | No | Ensembl | |
| rs782284863 | 145 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs782284863 | 145 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs2062572022 | 148 | A>V | No | Ensembl | |
| rs868991516 | 149 | S>P | No | Ensembl | |
| rs892706643 | 155 | S>L | No | gnomAD | |
| COSM3563491 | 156 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3096909 rs782731399 |
163 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2062572330 | 167 | K>E | No | TOPMed | |
| rs1557027117 | 170 | S>L | No | gnomAD | |
| rs373080668 | 171 | P>S | No |
ESP TOPMed gnomAD |
|
| rs373080668 | 171 | P>T | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 172 | R>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs375350173 | 172 | R>Q | No | gnomAD | |
| COSM3563492 | 173 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs782065899 | 174 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs1557027121 | 175 | Y>N | No | gnomAD | |
| rs2062572467 | 176 | R>M | No | TOPMed | |
|
rs782795164 TCGA novel |
178 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC TOPMed gnomAD NCI-TCGA |
|
COSM1718865 rs781914307 |
179 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs782506746 | 180 | H>Y | No |
ExAC gnomAD |
|
| rs952539577 | 182 | K>N | No | TOPMed | |
| rs1557027125 | 183 | F>I | No | gnomAD | |
| rs2062572697 | 184 | T>I | No | TOPMed | |
| rs1557027128 | 185 | H>D | No | gnomAD | |
| rs376606028 | 185 | H>Q | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 185 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1469427 | 187 | L>F | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2062572759 | 188 | P>R | No | TOPMed | |
| TCGA novel | 189 | R>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs782793816 | 190 | P>T | No |
ExAC gnomAD |
|
| rs781824473 | 192 | D>G | No |
ExAC gnomAD |
|
| COSM3424956 | 193 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 194 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4111129 | 195 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 197 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs982601422 | 197 | P>L | No | Ensembl | |
| rs1557027134 | 199 | M>I | No | gnomAD | |
| rs1557027133 | 199 | M>T | No | gnomAD | |
| rs782499063 | 200 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM6188130 | 201 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2062572912 | 201 | R>K | No |
TOPMed gnomAD |
|
| rs1557027137 | 203 | L>H | No | gnomAD | |
| rs782565867 | 205 | H>Q | No |
ExAC TOPMed gnomAD |
|
|
rs2062572977 RCV001355255 |
206 | Y>C | No |
ClinVar Ensembl dbSNP |
1 associated diseases with O43320
[MIM: 309630]: Metacarpal 4-5 fusion (MF4)
A rare congenital malformation of the hand characterized by the partial or complete fusion of the fourth and fifth metacarpals. The anomaly occurs as an isolated trait or part of a syndrome. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A rare congenital malformation of the hand characterized by the partial or complete fusion of the fourth and fifth metacarpals. The anomaly occurs as an isolated trait or part of a syndrome. . Note=The disease is caused by variants affecting the gene represented in this entry.
No regional properties for O43320
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for O43320 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11486 | FIBROBLAST GROWTH FACTOR |
| PANTHER Subfamily | PTHR11486:SF27 | FIBROBLAST GROWTH FACTOR 16 |
| PANTHER Protein Class |
intercellular signal molecule
growth factor |
|
| PANTHER Pathway Category |
FGF signaling pathway FGF |
|
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| extracellular space | That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| fibroblast growth factor receptor binding | Binding to a fibroblast growth factor receptor (FGFR). |
| growth factor activity | The function that stimulates a cell to grow or proliferate. Most growth factors have other actions besides the induction of cell growth or proliferation. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| animal organ morphogenesis | Morphogenesis of an animal organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process in which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions. |
| cell differentiation | The cellular developmental process in which a relatively unspecialized cell, e.g. embryonic or regenerative cell, acquires specialized structural and/or functional features that characterize a specific cell. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell-cell signaling | Any process that mediates the transfer of information from one cell to another. This process includes signal transduction in the receiving cell and, where applicable, release of a ligand and any processes that actively facilitate its transport and presentation to the receiving cell. Examples include signaling via soluble ligands, via cell adhesion molecules and via gap junctions. |
| fibroblast growth factor receptor signaling pathway | The series of molecular signals generated as a consequence of a fibroblast growth factor receptor binding to one of its physiological ligands. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of endothelial cell chemotaxis to fibroblast growth factor | Any process that activates or increases the frequency, rate or extent of endothelial cell chemotaxis to fibroblast growth factor. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| response to temperature stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a temperature stimulus. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
28 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P48801 | FGF3 | Fibroblast growth factor 3 | Gallus gallus (Chicken) | SS |
| O15520 | FGF10 | Fibroblast growth factor 10 | Homo sapiens (Human) | SS |
| P10767 | FGF6 | Fibroblast growth factor 6 | Homo sapiens (Human) | SS |
| P11487 | FGF3 | Fibroblast growth factor 3 | Homo sapiens (Human) | SS |
| P31371 | FGF9 | Fibroblast growth factor 9 | Homo sapiens (Human) | EV |
| Q92914 | FGF11 | Fibroblast growth factor 11 | Homo sapiens (Human) | SS |
| Q9HCT0 | FGF22 | Fibroblast growth factor 22 | Homo sapiens (Human) | SS |
| Q9NP95 | FGF20 | Fibroblast growth factor 20 | Homo sapiens (Human) | SS |
| Q9NSA1 | FGF21 | Fibroblast growth factor 21 | Homo sapiens (Human) | PR |
| P08620 | FGF4 | Fibroblast growth factor 4 | Homo sapiens (Human) | PR |
| P21658 | Fgf6 | Fibroblast growth factor 6 | Mus musculus (Mouse) | SS |
| Q9ESS2 | Fgf22 | Fibroblast growth factor 22 | Mus musculus (Mouse) | SS |
| O35565 | Fgf10 | Fibroblast growth factor 10 | Mus musculus (Mouse) | SS |
| P54130 | Fgf9 | Fibroblast growth factor 9 | Mus musculus (Mouse) | SS |
| Q9JJN1 | Fgf21 | Fibroblast growth factor 21 | Mus musculus (Mouse) | PR |
| Q9ESL9 | Fgf20 | Fibroblast growth factor 20 | Mus musculus (Mouse) | SS |
| P11403 | Fgf4 | Fibroblast growth factor 4 | Mus musculus (Mouse) | PR |
| P61329 | Fgf12 | Fibroblast growth factor 12 | Mus musculus (Mouse) | PR |
| P05524 | Fgf3 | Fibroblast growth factor 3 | Mus musculus (Mouse) | SS |
| Q9ESL8 | Fgf16 | Fibroblast growth factor 16 | Mus musculus (Mouse) | SS |
| Q95L12 | FGF9 | Fibroblast growth factor 9 | Sus scrofa (Pig) | SS |
| Q9EST9 | Fgf20 | Fibroblast growth factor 20 | Rattus norvegicus (Rat) | SS |
| P36364 | Fgf9 | Fibroblast growth factor 9 | Rattus norvegicus (Rat) | SS |
| P70492 | Fgf10 | Fibroblast growth factor 10 | Rattus norvegicus (Rat) | SS |
| O54769 | Fgf16 | Fibroblast growth factor 16 | Rattus norvegicus (Rat) | SS |
| Q6PBT8 | fgf1 | Putative fibroblast growth factor 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| P48802 | fgf3 | Fibroblast growth factor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q2HXK8 | fgf16 | Fibroblast growth factor 16 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAEVGGVFAS | LDWDLHGFSS | SLGNVPLADS | PGFLNERLGQ | IEGKLQRGSP | TDFAHLKGIL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RRRQLYCRTG | FHLEIFPNGT | VHGTRHDHSR | FGILEFISLA | VGLISIRGVD | SGLYLGMNER |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GELYGSKKLT | RECVFREQFE | ENWYNTYAST | LYKHSDSERQ | YYVALNKDGS | PREGYRTKRH |
| 190 | 200 | ||||
| QKFTHFLPRP | VDPSKLPSMS | RDLFHYR |